Systematic, Genome-Wide Identification of Host Genes Affecting Replication of a Positive-Strand RNA Virus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Detection of Infectious Brome Mosaic Virus in Irrigation Ditches and Draining Strands in Poland
Eur J Plant Pathol https://doi.org/10.1007/s10658-018-1531-7 Detection of infectious Brome mosaic virus in irrigation ditches and draining strands in Poland Małgorzata Jeżewska & Katarzyna Trzmiel & Aleksandra Zarzyńska-Nowak Accepted: 29 June 2018 # The Author(s) 2018 Abstract Environmental waters, e.g. rivers, lakes Results confirmed the highest amino acid sequence and irrigation water, are a good source of many homology in the fragment of polymerase 2a (99.2% plant viruses. The pathogens can infect plants get- – 100%) and the most divergence in CP (96.2% - ting through damaged root hairs or small wounds 100%). This is the first report on the detection of an that appear during plant growth. First results dem- infective cereal virus in aqueous environment. onstrated common incidence of Tobacco mosaic virus (TMV) and Tomato mosaic virus (ToMV) in Keywords BMV. Water-borne virus . Cereals . RT-PCR water samples collected from irrigation ditches and drainage canals surrounding fields in Southern Greater Poland. Principal objective of this work The occurrence of plant viruses in aqueous environment was to examine if environmental water might be was studied less intensively than other water-borne vi- the source of viruses infective to cereals. The in- ruses having impact on human health. Mehle and vestigation was focused on mechanically transmit- Ravnikar (2012) thoroughly reviewed the reports and ted pathogens. Virus identification was performed listed 16 plant virus species isolated from different water by biological, electron microscopic, serological and sources, mainly from Europe, but not from Poland. molecular methods. Preliminary assays demonstrat- The main objective of our work was to fulfil this gap ed Bromemosaicvirus(BMV) infections in symp- with special attention focused on infective cereal viruses. -

The Crystallographic Structure of Brome Mosaic Virus Robertw.Lucas,Stevenb.Larsonandalexandermcpherson*
doi:10.1006/jmbi.2001.5389availableonlineathttp://www.idealibrary.comon J. Mol. Biol. (2002) 317, 95±108 The Crystallographic Structure of Brome Mosaic Virus RobertW.Lucas,StevenB.LarsonandAlexanderMcPherson* University of California, Irvine The structure of brome mosaic virus (BMV), the type member of the 560 Steinhaus Hall, Irvine bromoviridae family, has been determined from a single rhombohedral CA 92697-3900, USA crystal by X-ray diffraction, and re®ned to an R value of 0.237 for data in the range 3.4-40.0 AÊ . The structure, which represents the native, compact form at pH 5.2 in the presence of 0.1 M Mg2, was solved by molecular replacement using the model of cowpea chlorotic mottle virus (CCMV), which BMV closely resembles. The BMV model contains amino acid resi- dues 41-189 for the pentameric capsid A subunits, and residues 25-189 and 1-189 for the B and C subunits, respectively, which compose the hex- americ capsomeres. In the model there are two Mg ions and one mol- ecule of polyethylene glycol (PEG). The ®rst 25 amino acid residues of the C subunit are modeled as polyalanine. The coat protein has the cano- nical ``jellyroll'' b-barrel topology with extended amino-terminal polypep- tides as seen in other icosahedral plant viruses. Mass spectrometry shows that in native BMV virions, a signi®cant fraction of the amino-terminal peptides are apparently cleaved. No recognizable nucleic acid residue is visible in the electron density maps except at low resolution where it appears to exhibit a layered arrangement in the virion interior. -

Tically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 21 June 2021 doi:10.20944/preprints202106.0526.v1 Review Towards the forest virome: next-generation-sequencing dras- tically expands our understanding on virosphere in temperate forest ecosystems Artemis Rumbou 1,*, Eeva J. Vainio 2 and Carmen Büttner 1 1 Faculty of Life Sciences, Albrecht Daniel Thaer-Institute of Agricultural and Horticultural Sciences, Humboldt-Universität zu Berlin, Ber- lin, Germany; [email protected], [email protected] 2 Natural Resources Institute Finland, Latokartanonkaari 9, 00790, Helsinki, Finland; [email protected] * Correspondence: [email protected] Abstract: Forest health is dependent on the variability of microorganisms interacting with the host tree/holobiont. Symbiotic mi- crobiota and pathogens engage in a permanent interplay, which influences the host. Thanks to the development of NGS technol- ogies, a vast amount of genetic information on the virosphere of temperate forests has been gained the last seven years. To estimate the qualitative/quantitative impact of NGS in forest virology, we have summarized viruses affecting major tree/shrub species and their fungal associates, including fungal plant pathogens, mutualists and saprotrophs. The contribution of NGS methods is ex- tremely significant for forest virology. Reviewed data about viral presence in holobionts, allowed us to address the role of the virome in the holobionts. Genetic variation is a crucial aspect in hologenome, significantly reinforced by horizontal gene transfer among all interacting actors. Through virus-virus interplays synergistic or antagonistic relations may evolve, which may drasti- cally affect the health of the holobiont. Novel insights of these interplays may allow practical applications for forest plant protec- tion based on endophytes and mycovirus biocontrol agents. -

Synthesis and Characterization of a Full-Length Infectious Cdna Clone of Tomato Mottle Mosaic Virus
viruses Article Synthesis and Characterization of a Full-Length Infectious cDNA Clone of Tomato Mottle Mosaic Virus Liqin Tu 1,2 , Shuhua Wu 2, Danna Gao 1, Yong Liu 3, Yuelin Zhu 1,* and Yinghua Ji 2,* 1 College of Horticulture, Nanjing Agricultural University, Nanjing 210095, China; [email protected] (L.T.); [email protected] (D.G.) 2 Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences/Key Lab of Food Quality and Safety of Jiangsu Province-State Key Laboratory Breeding Base, Nanjing 210014, China; [email protected] 3 Institute of Plant Protection, Hunan Academy of Agricultural Sciences, Changsha 410125, China; [email protected] * Correspondence: [email protected] (Y.Z.); [email protected] (Y.J.); Tel.: +86-25-84396472 (Y.Z.); +86-25-84390394 (Y.J.) Abstract: Tomato mottle mosaic virus (ToMMV) is a noteworthy virus which belongs to the Virgaviridae family and causes serious economic losses in tomato. Here, we isolated and cloned the full-length genome of a ToMMV Chinese isolate (ToMMV-LN) from a naturally infected tomato (Solanum lycopersicum L.). Sequence analysis showed that ToMMV-LN contains 6399 nucleotides (nts) and is most closely related to a ToMMV Mexican isolate with a sequence identity of 99.48%. Next, an infectious cDNA clone of ToMMV was constructed by a homologous recombination approach. Both the model host N. benthamiana and the natural hosts tomato and pepper developed severe symptoms upon agroinfiltration with pToMMV, which had a strong infectivity. Electron micrographs indicated that a large number of rigid rod-shaped ToMMV virions were observed from the agroinfiltrated N. -

Crystallization of Brome Mosaic Virus and T = 1 Brome Mosaic
Virology 286, 290–303 (2001) doi:10.1006/viro.2000.0897, available online at http://www.idealibrary.com on Crystallization of Brome Mosaic Virus and T ϭ 1 Brome Mosaic Virus Particles Following a Structural Transition Robert W. Lucas, Yurii G. Kuznetsov, Steven B. Larson, and Alexander McPherson1 University of California, Irvine, Department of Molecular Biology and Biochemistry, Irvine, California 92697-3900 Received November 9, 2000; returned to author for revision January 17, 2001; accepted March 6, 2001 Brome mosaic virus (BMV), a T ϭ 3 icosahedral plant virus, can be dissociated into coat protein subunits and subunit oligomers at pH 7.5 in the presence of concentrated salts. We have found that during the course of this treatment the coat protein subunits are cleaved, presumably by plant cell proteases still present in the preparation, between amino acids 35 and 36. The truncated protein subunits will then reorganize into T ϭ 1 icosahedral particles and can be crystallized from sodium malonate. Quasi elastic light scattering and atomic force microscopy results suggest that the transition from T ϭ 3toT ϭ 1 particles can occur by separate pathways, dissociation into coat protein subunits and oligomers and reassembly into T ϭ 1 particles, or direct condensation of the T ϭ 3 virions to T ϭ 1 particles with the shedding of hexameric capsomeres. The latter process has been directly visualized using atomic force microscopy. Native T ϭ 3 virions have been crystallized in several different crystal forms, but neither a rhombohedral form nor either of two orthorhombic forms diffract beyond about 3.4 Å. -
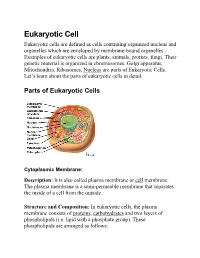
Eukaryotic Cell Eukaryotic Cells Are Defined As Cells Containing Organized Nucleus and Organelles Which Are Enveloped by Membrane-Bound Organelles
Eukaryotic Cell Eukaryotic cells are defined as cells containing organized nucleus and organelles which are enveloped by membrane-bound organelles. Examples of eukaryotic cells are plants, animals, protists, fungi. Their genetic material is organized in chromosomes. Golgi apparatus, Mitochondria, Ribosomes, Nucleus are parts of Eukaryotic Cells. Let’s learn about the parts of eukaryotic cells in detail. Parts ot Eukaryotic Cells Cytoplasmic Membrane: Description: It is also called plasma membrane or cell membrane. The plasma membrane is a semi-permeable membrane that separates the inside of a cell from the outside. Structure and Composition: In eukaryotic cells, the plasma membrane consists of proteins , carbohydrates and two layers of phospholipids (i.e. lipid with a phosphate group). These phospholipids are arranged as follows: • The polar, hydrophilic (water-loving) heads face the outside and inside of the cell. These heads interact with the aqueous environment outside and within a cell. • The non-polar, hydrophobic (water-repelling) tails are sandwiched between the heads and are protected from the aqueous environments. Scientists Singer and Nicolson(1972) described the structure of the phospholipid bilayer as the ‘Fluid Mosaic Model’. The reason is that the bi-layer looks like a mosaic and has a semi-fluid nature that allows lateral movement of proteins within the bilayer. Image: Fluid mosaic model. Orange circles – Hydrophilic heads; Lines below – Hydrophobic tails. Functions • The plasma membrane is selectively permeable i.e. it allows only selected substances to pass through. • It protects the cells from shock and injuries. • The fluid nature of the membrane allows the interaction of molecules within the membrane. -

Positive-Strand RNA Viruses Stimulate Host Phosphatidylcholine Synthesis at Viral Replication Sites
Positive-strand RNA viruses stimulate host phosphatidylcholine synthesis at viral replication sites Jiantao Zhanga,1, Zhenlu Zhanga,b, Vineela Chukkapallic, Jules A. Nchoutmboubed, Jianhui Lia, Glenn Randallc, George A. Belovd, and Xiaofeng Wanga,2 aDepartment of Plant Pathology, Physiology, and Weed Science, Virginia Tech, Blacksburg, VA 24061; bDepartment of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou 350002, People’s Republic of China; cDepartment of Microbiology, The University of Chicago, Chicago, IL 60637; and dVirginia-Maryland Regional College of Veterinary Medicine, University of Maryland, College Park, MD 20742 Edited by Peter Palese, Icahn School of Medicine at Mount Sinai, New York, NY, and approved January 12, 2016 (received for review October 6, 2015) All positive-strand RNA viruses reorganize host intracellular mem- expression of 1a alone in yeast induces spherule formation (6), branes to assemble their viral replication complexes (VRCs); how- which requires 1a’s amphipathic α-helix (1a amino acids 392–407) ever, how these viruses modulate host lipid metabolism to (8), helix A, and 1a–1a interactions (9, 10). In addition, several accommodate such membrane proliferation and rearrangements is host proteins, including membrane-shaping reticulons (RTNs) not well defined. We show that a significantly increased phospha- (11) and an ESCRT (endosomal sorting complex required for tidylcholine (PC) content is associated with brome mosaic virus transport) component, Snf7p (sucrose nonfermenting7) (12), are (BMV) replication in both natural host barley and alternate host recruited by 1a to form spherules. yeast based on a lipidomic analysis. Enhanced PC levels are primarily Similar to other (+)RNA viruses, BMV promotes host lipid associated with the perinuclear ER membrane, where BMV replica- synthesis and requires balanced lipids for the formation and ac- tion takes place. -

Trna Elements Mediate the Assembly of an Icosahedral RNA Virus
tRNA elements mediate the assembly of an icosahedral RNA virus Yoon Gi Choi*, Theo W. Dreher†, and A. L. N. Rao*‡ *Department of Plant Pathology, University of California, Riverside, CA, 92521-0122; and †Department of Microbiology, Oregon State University, Corvallis, OR 97331-3804 Communicated by George Bruening, University of California, Davis, CA, November 20, 2001 (received for review August 21, 2001) tRNAs, the adapter molecules in protein synthesis, also serve as signals necessary for minus-strand initiation and synthesis by the metabolic cofactors and as primers for viral RNA-directed DNA syn- BMV replicase (5, 13). thesis. The genomic and subgenomic RNAs of some plant viruses have BMV coat protein (CP) is composed of 189 amino acids and -a3-terminal tRNA-like structure (TLS) that can accept a specific amino assembles into mature icosahedral virions with T ϭ 3 quasisym acid and serve as a site for initiation of replication and as a simple metry (9, 14). Purified RNA and CP subunits can be reassembled telomere. We report a previously undescribed role for the TLS of in vitro to produce infectious particles indistinguishable from those brome mosaic virus (BMV), and potentially for cellular tRNA, in assembled in vivo (9, 15). Although empty capsids can assemble in mediating the assembly of its icosahedral virions. BMV genomic RNAs vitro at low pH (Ϸ5), they do not form in vitro under physiological and subgenomic RNA lacking the TLS failed to assemble into virions conditions of low salt and neutral pH and are not observed in vivo when incubated with purified BMV coat protein. -

Alphavirus-Induced Membrane Rearrangements During Replication, Assembly, and Budding
pathogens Review Alphavirus-Induced Membrane Rearrangements during Replication, Assembly, and Budding Zeinab Elmasri 1,2, Benjamin L. Nasal 2 and Joyce Jose 1,2,* 1 Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA 16802, USA; [email protected] 2 Department of Biochemistry & Molecular Biology, Eberly College of Science, The Pennsylvania State University, University Park, PA 16802, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-814-863-8806 Abstract: Alphaviruses are arthropod-borne viruses mainly transmitted by hematophagous insects that cause moderate to fatal disease in humans and other animals. Currently, there are no approved vaccines or antivirals to mitigate alphavirus infections. In this review, we summarize the current knowledge of alphavirus-induced structures and their functions in infected cells. Throughout their lifecycle, alphaviruses induce several structural modifications, including replication spherules, type I and type II cytopathic vacuoles, and filopodial extensions. Type I cytopathic vacuoles are replication-induced structures containing replication spherules that are sites of RNA replication on the endosomal and lysosomal limiting membrane. Type II cytopathic vacuoles are assembly induced structures that originate from the Golgi apparatus. Filopodial extensions are induced at the plasma membrane and are involved in budding and cell-to-cell transport of virions. This review provides an overview of the viral and host factors involved in the biogenesis and function of these virus-induced structures. Understanding virus–host interactions in infected cells will lead to the identification of Citation: Elmasri, Z.; Nasal, B.L.; new targets for antiviral discovery. Jose, J. Alphavirus-Induced Membrane Rearrangements during Keywords: Togaviridae; alphavirus; spherule; replication; cytopathic vacuole; nucleocapsid core; Replication, Assembly, and Budding. -

Determination of Protein Interactions Among Replication Components of Apple Necrotic Mosaic Virus
viruses Article Determination of Protein Interactions among Replication Components of Apple Necrotic Mosaic Virus Zhen-Lu Zhang, Fu-Jun Zhang, Peng-Fei Zheng, Yin-Huan Xie, Chun-Xiang You and Yu-Jin Hao * State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an 271000, China; [email protected] (Z.-L.Z.); [email protected] (F.-J.Z.); [email protected] (P.-F.Z.); [email protected] (Y.-H.X.); [email protected] (C.-X.Y.) * Correspondence: [email protected] Received: 16 February 2020; Accepted: 20 April 2020; Published: 22 April 2020 Abstract: Apple mosaic disease is one of the most widely distributed and destructive diseases in apple cultivation worldwide, especially in China, whose apple yields account for more than 50% of the global total. Apple necrotic mosaic virus (ApNMV) is a newly identified ilarvirus that is closely associated with apple mosaic disease in China; however, basic viral protein interactions that play key roles in virus replication and the viral life cycle have not been determined in ApNMV. Here, we first identify an ApNMV–Lw isolate that belongs to subgroup 3 in the genus Ilarvirus. ApNMV–Lw was used to investigate interactions among viral components. ApNMV 1a and 2apol, encoded by RNA1 and RNA2, respectively, were co-localized in plant cell cytoplasm. ApNMV 1a interacted with itself at both the inter- and intramolecular levels, and its N-terminal portion played a key role in these interactions. 1a also interacted with 2apol, and 1a’s C-terminal, together with 2apol’s N-terminal, was required for this interaction. -

JOZEF JULIAN BUJARSKI, Professor of Biological Sciences
CURRICULUM VITAE JOZEF JULIAN BUJARSKI Professor of Biological Sciences Northern Illinois University Department of Biological Sciences DeKalb, IL 60115 DATE: January, 2015. EDUCATION M.S., Adam Mickiewicz University, Bioorganic Chemistry, Poznan, Poland, 1972 M.S., Adam Mickiewicz University, Molecular Biology/Biochemistry, Poznan, Poland, 1976 Ph.D., Adam Mickiewicz University, Bioorganic Chemistry, Poznan, Poland, 1978 D.Sc. (Doctor of Science), Institute of Bioorganic Chemistry, Polish Academy of Sciences, May 1998 AREAS OF SPECIALIZATION RNA viruses; Molecular Biology; Biotechnology; Nucleic Acids PROFESSIONAL EXPERIENCE Director of Plant Molecular Biology Center at Northern Illinois University, 2004-present State Professor of the Republic of Poland, July 2003-present Distinguished Research Professor, Northern Illinois University, 2001-present. Professor, Northern Illinois University, Department of Biological Sciences, 1995-present Associate Professor, Northern Illinois University, Department of Biological Sciences, 1991-1994 Assistant Professor, Northern Illinois University, Department of Biological Sciences, 1987-1991 Assistant Scientist, University of Wisconsin-Madison, Biophysics Laboratory, 1985-1987 Research Associate, University of Wisconsin-Madison, Department of Horticulture, 1981-1984 Senior Scientist (Adjunct), Polish Academy of Sciences, Institute of Plant Genetics, Poznan, Poland, 1978-1981 Research Associate, A. Mickiewicz University, Department of Biology, Poznan, Poland, 1976-1978 Research Assistant, A. Mickiewicz University, Department of Chemistry, Poznan, Poland, 1972-1976 Research Assistant, A. Mickiewicz University, Department of Chemistry, Poznan, Poland 1970-1971 HONORS AND AWARDS Ph.D. Graduate Fellowship to Adam Mickiewicz University, Poznan, Poland, 1972-1976. Graduated with honors in the class of bioorganic chemistry, 1972. Tenure at NIU/BIOS, 1991. Presidential Research Professorship Award, Northern Illinois University, 1997. State Professorship from the President of the Republic of Poland, 2003. -

Towards the Forest Virome: High-Throughput Sequencing Drastically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems
microorganisms Review Towards the Forest Virome: High-Throughput Sequencing Drastically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems Artemis Rumbou 1,* , Eeva J. Vainio 2 and Carmen Büttner 1 1 Faculty of Life Sciences, Albrecht Daniel Thaer-Institute of Agricultural and Horticultural Sciences, Humboldt-Universität zu Berlin, 14195 Berlin, Germany; [email protected] 2 Natural Resources Institute Finland, Forest Health and Biodiversity, Latokartanonkaari 9, 00790 Helsinki, Finland; eeva.vainio@luke.fi * Correspondence: [email protected] Abstract: Thanks to the development of HTS technologies, a vast amount of genetic information on the virosphere of temperate forests has been gained in the last seven years. To estimate the qualitative/quantitative impact of HTS on forest virology, we have summarized viruses affecting major tree/shrub species and their fungal associates, including fungal plant pathogens, mutualists and saprotrophs. The contribution of HTS methods is extremely significant for forest virology. Reviewed data on viral presence in holobionts allowed us a first attempt to address the role of virome in holobionts. Forest health is dependent on the variability of microorganisms interacting with the host tree/holobiont; symbiotic microbiota and pathogens engage in a permanent interplay, which influences the host. Through virus–virus interplays synergistic or antagonistic relations may evolve, which may drastically affect the health of the holobiont. Novel insights of these interplays may allow Citation: Rumbou, A.; Vainio, E.J.; Büttner, C. Towards the Forest practical applications for forest plant protection based on endophytes and mycovirus biocontrol Virome: High-Throughput agents. The current analysis is conceived in light of the prospect that novel viruses may initiate an Sequencing Drastically Expands Our emergent infectious disease and that measures for the avoidance of future outbreaks in forests should Understanding on Virosphere in be considered.