Final Dissertation Front Matter
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Elements by Townrange for Racine County
Elements by Townrange for Racine County The Natural Heritage Inventory (NHI) database contains recent and historic element (rare species and natural community) observations. A generalized version of the NHI database is provided below as a general reference and should not be used as a substitute for a WI Dept of Natural Resources NHI review of a specific project area. The NHI database is dynamic, records are continually being added and/or updated. The following data are current as of 03/26/2014: Town Range State Federal State Global Group Scientific Name Common Name Status Status Rank Rank Name Agalinis auriculata Earleaf Foxglove SC S1 G3 Plant Asclepias lanuginosa Woolly Milkweed THR S1 G4? Plant Asclepias purpurascens Purple Milkweed END S3 G5? Plant Asclepias sullivantii Prairie Milkweed THR S2S3 G5 Plant Carex garberi Elk Sedge THR S2 G5 Plant~ Carex swanii Swan Sedge SC S1 G5 Plant Cypripedium candidum Small White Lady's-slipper THR S3 G4 Plant~ Dryopteris clintoniana Clinton's Woodfern SC SH G5 Plant Lespedeza leptostachya Prairie Bush-clover END LT S2 G3 Plant Phegopteris hexagonoptera Broad Beech Fern SC S2 G5 Plant Plantago cordata Heart-leaved Plantain END S1 G4 Plant~ Prenanthes aspera Rough Rattlesnake-root END S1 G4? Plant Ranunculus cymbalaria Seaside Crowfoot THR S2 G5 Plant~ Thamnophis proximus Western Ribbonsnake END S1 G5 Snake~ Thamnophis sauritus Eastern Ribbonsnake END S1 G5 Snake~ 001N019E Moxostoma carinatum River Redhorse THR S2 G4 Fish~ 001N020E Moxostoma carinatum River Redhorse THR S2 G4 Fish~ 001N022E Aphredoderus -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Recovering After Childbirth in the Mixtec Highlands (Mexico)
M~DICAMENTSET ALIMENTS :L'APPROCHE ETHNOPHARMACOLOGIQLJE I 99 Recovering after childbirth in the Mixtec highlands (Mexico) KATZ Esther ORSTOM (Institut Français de Recherche Scientifique pourle Développement en Coopération) Département MAA (Milieu et Activités Agricoles) 213, rue Lafayette- 75480 PARIS Cedex 10 - FRANCE Fa: 33-1-40351713 RÉSUMÉ Les Indiens du haut pays mixtèque, tout comme d'autres Indiens du Mexique, prennent particulièrement soin des jeunes accouchées. Un certain nombre de travaux portent sur la grossesse et l'accouchement au Mexique, mais le thème du post- partum a été peu étudié en profondeur, bien que les indigknes insistent sur le danger et l'importance des soins à cette période. Dans ce travail, la conception, la grossesse et l'accouchement sont décrits à titre introductif, tandis que les pratiques du post- partum sont analysées en détail : la période de repos de 20 ou 40 jours, le régime alimentaire particulier, l'abstinence sexuelle, les diverses précautions et prohibitions, les soins corporels, les tisanes, les bains de plantes et surtout, le bain de vapeur, à fonction à la fois thérapeutique et rituelle. L'article posele problème de l'analyse des données touchant aux pratiques corpo- relles féminines, difficilement verbalisées. Il amorce également une comparaison avec les pratiques des pays industrialisés occidentaux et suggère de puiser dans les pratiques et les connaissances des sociétés dites << traditionnelles >> pour remédier aux dépressions post-partum. INTRODUCTION 1973;ALVAREZ JEYLlENREICH, 1976; COMINSKY, 1976 1982; QUEZADA 1977;RITA, 1979;L6PEZAUSTIN, Like most of the indigenous peoples of Mexico, the Mixtec AND 1980; GARCfARUIZANDPETRICH, 1983; IRETON, 1987; Indians view pregnancyas a disease. -

Chromosome Numbers in Compositae, XII: Heliantheae
SMITHSONIAN CONTRIBUTIONS TO BOTANY 0 NCTMBER 52 Chromosome Numbers in Compositae, XII: Heliantheae Harold Robinson, A. Michael Powell, Robert M. King, andJames F. Weedin SMITHSONIAN INSTITUTION PRESS City of Washington 1981 ABSTRACT Robinson, Harold, A. Michael Powell, Robert M. King, and James F. Weedin. Chromosome Numbers in Compositae, XII: Heliantheae. Smithsonian Contri- butions to Botany, number 52, 28 pages, 3 tables, 1981.-Chromosome reports are provided for 145 populations, including first reports for 33 species and three genera, Garcilassa, Riencourtia, and Helianthopsis. Chromosome numbers are arranged according to Robinson’s recently broadened concept of the Heliantheae, with citations for 212 of the ca. 265 genera and 32 of the 35 subtribes. Diverse elements, including the Ambrosieae, typical Heliantheae, most Helenieae, the Tegeteae, and genera such as Arnica from the Senecioneae, are seen to share a specialized cytological history involving polyploid ancestry. The authors disagree with one another regarding the point at which such polyploidy occurred and on whether subtribes lacking higher numbers, such as the Galinsoginae, share the polyploid ancestry. Numerous examples of aneuploid decrease, secondary polyploidy, and some secondary aneuploid decreases are cited. The Marshalliinae are considered remote from other subtribes and close to the Inuleae. Evidence from related tribes favors an ultimate base of X = 10 for the Heliantheae and at least the subfamily As teroideae. OFFICIALPUBLICATION DATE is handstamped in a limited number of initial copies and is recorded in the Institution’s annual report, Smithsonian Year. SERIESCOVER DESIGN: Leaf clearing from the katsura tree Cercidiphyllumjaponicum Siebold and Zuccarini. Library of Congress Cataloging in Publication Data Main entry under title: Chromosome numbers in Compositae, XII. -

Lafayette Creek Property—Phases I and Ii Umbrella
LAFAYETTE CREEK PROPERTY—PHASES I AND II UMBRELLA REGIONAL MITIGATION PLANS FOR FLORIDA DEPARTMENT OF TRANSPORTATION PROJECTS CONCEPTUAL MITIGATION PLAN WALTON COUNTY, FLORIDA June 20, 2011 Prepared for: Mr. David Clayton Northwest Florida Water Management District 81 Water Management Drive Havana, FL 32333 Prepared by: ________________________________ ________________________________ Caitlin E. Elam Richard W. Cantrell Staff Scientist Senior Consultant 4240-034 Y100 Lafayette Creek Phases I and II Restoration Plan 062011_E.doc Lafayette Creek Property—Phases I and II Umbrella Regional Mitigation Plans for Florida Department of Transportation Projects June 20, 2011 TABLE OF CONTENTS 1.0 PROJECT OVERVIEW AND GOALS ............................................................................. 1 2.0 LOCATION AND LANDSCAPE ...................................................................................... 2 3.0 EXISTING CONDITIONS ................................................................................................. 2 4.0 LISTED SPECIES ............................................................................................................ 13 5.0 EXOTIC SPECIES ............................................................................................................ 17 6.0 HISTORIC CONDITIONS ............................................................................................... 17 7.0 SOILS ............................................................................................................................... -

CDLT Mountain Home & USFS-Boundry Butte Plant List
CDLT Mountain Home Preserve- Boundary Butte Plant list CDLT Mountain Home & USFS-Boundry Butte Plant list Type Scientific Name Common Name Fern Pteridium aquilinum bracken fern Forb Achillea millefolium common yarrow Forb Agoseris heterophylla annual agoseris Forb Anemone oregana Oregon anemone Forb Antennaria racemosa raceme pussytoes Forb Boechera pauciflorus rockcress (Formerly Arabis) Forb Arnica cordifolia heart-leaf arnica Forb Balsamorhiza sagittata arrowleaf balsamroot Forb Brickellia oblongifolia Mojave brickellbush Forb Cacaliopsis nardosmia silvercrown (Formerly Luina) Forb Calochortus lyallii Lyall's mariposa lily Forb Camassia quamash common camas Forb Castilleja miniata scarlet Indian paintbrush Forb Claytonia lanceolata springbeauty Forb Collinsia parviflora small-flowered blue-eyed mary Forb Commandra umbellata bastard toadflax Forb Delphinium viridescens Wenatchee larkspur Forb Erythronium grandiflorum glacier lily Forb Erysimum species wallflower Forb Fragaria virginiana Virginia strawberry Forb Fritillaria affinis checker lily, chocolate lily Forb Fritillaria pudica yellow bells Forb Galium sp. bedstraw Forb Heuchera cylindrica roundleaf alumroot Forb Hydrophyllum capitatum ballhead waterleaf Forb Lathyrus pauciflorus few-flowered pea Forb Lithophragma parviflorum small-flowered woodland-star Forb Lithophragma glabrum bulbous woodland-star Forb Lithophragma tenellum slender woodland-star Forb Lomatium nudicaule barestem biscuitroot Forb Lomatium triternatum nineleaf biscuitroot Forb Lonicera ciliosa orange honeysuckle -
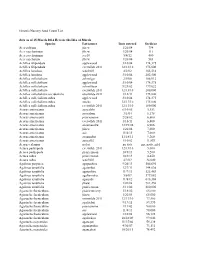
Q Seed Counts
Genesis Nursery Seed Count List data as of 15 March 2014 Beware ths Ides of March Species Lot/source Date entered Seeds/oz Acer rubrum jfnew 1/26/04 794 Acer saccharinum jfnew 1/26/04 111 Acer saccharinum aes10 5/4/12 400 Acer saccharum jfnew 1/26/04 563 Achillea filipendula applewood 3/10/04 174,375 Achillea filipendula everwilde 2011 12/13/10 175,000 Achillea lanulosa wns2001 4/2/02 165,516 Achillea lanulosa applewood 3/10/04 202,500 Achillea millefoilium achmilgo 2/9/06 166,912 Achillea millefoilium applewood 3/10/04 174,375 Achillea millefoilium achmil0san 9/21/02 179,022 Achillea millefoilium everwilde 2011 12/13/10 200,000 Achillea millefoilium occidentalis everwilde 2011 1/16/11 175,000 Achillea millefoilium rubra applewood 3/10/04 174,375 Achillea millefoilium rubra stocks 12/13/10 175,000 Achillea millefoilium rubra everwilde 2011 12/13/10 180,000 Acorus americanus acocalshi 6/10/05 5,553 Acorus americanus acocalsan 3/1/04 6,175 Acorus americanus prairiemoon 2/26/02 6,600 Acorus americanus everwilde 2011 1/16/11 6,800 Acorus americanus acoameroku 12/19/06 6,906 Acorus americanus jfnew 1/26/04 7,000 Acorus americanus aes 11/4/11 7,000 Acorus americanus acoamebat 11/18/11 9,260 Acorus americanus acocal02 1/10/02 11,853 Acorus calamus no lot no date zip, nada, zilch Actaea pachypoda everwilde 2011 12/13/10 5,000 Actaea pachypoda prairiemoon 10/3/13 5,200 Actaea rubra prairiemoon 10/3/13 4,450 Actaea rubra wns2001 4/2/02 34,000 Agalinus purpurea agapuuhiru 9/26/13 506,696 Agalinus tenuifolia agatenbat 12/7/11 144,036 Agalinus tenuifolia -

La Flora Patrimonial De Quito Descubierta Por La Expedición De
AVANCES EN CIENCIAS E INGENIERÍAS ARTÍCULO/ARTICLE SECCIÓN/SECTION B La flora patrimonial de Quito descubierta por la expedición de Humboldt y Bonpland en el año 1802 Carlos Ruales1,∗ Juan E. Guevara2 1Universidad San Francisco de Quito, Colegio de Agricultura, Alimentos y Nutrición. Diego de Robles y Vía Interoceánica, Quito, Ecuador. 2Herbario Alfredo Paredes (QAP), Facultad de Filosofía y Letras, Universidad Central del Ecuador Apartado Postal 17-01-2177, Quito, Ecuador. ∗Autor principal/Corresponding author, e-mail: [email protected] Editado por/Edited by: D. F. Cisneros-Heredia, M.Sc. Recibido/Received: 04/14/2010. Aceptado/Accepted: 08/25/2010. Publicado en línea/Published on Web: 12/08/2010. Impreso/Printed: 12/08/2010. Abstract The results from this research are based on historical data and data from herbarium co- llections prepared by Alexander von Humboldt and Aimé Bonpland in 1802 in the city of Quito and its surroundings. In the research, 142 species from the Humboldt and Bonpland’s collections have been selected because of their patrimonial value for the city’s inhabitants; those species were collected for the first time in this area and the plant collections include type-specimens used to describe the species. Twenty-five species are endemic to Ecuador and from those, Cynanchum serpyllifolium (Asclepiadaceae) has not been found for more than 100 years, while Aetheolaena ledifolia (Asteraceae) and Cyperus multifolius (Cypera- ceae) are only known from their type-collections made in 1802. Almost 80 per cent of the collected species were herbs, which showed the advanced human intervention at the time. These premises help to propose a plan that manages patrimonial plant concepts in Quito and its surrounding towns and to increase the appropriation and valorization processes. -

The Vascular Flora of Rarău Massif (Eastern Carpathians, Romania). Note Ii
Memoirs of the Scientific Sections of the Romanian Academy Tome XXXVI, 2013 BIOLOGY THE VASCULAR FLORA OF RARĂU MASSIF (EASTERN CARPATHIANS, ROMANIA). NOTE II ADRIAN OPREA1 and CULIŢĂ SÎRBU2 1 “Anastasie Fătu” Botanical Garden, Str. Dumbrava Roşie, nr. 7-9, 700522–Iaşi, Romania 2 University of Agricultural Sciences and Veterinary Medicine Iaşi, Faculty of Agriculture, Str. Mihail Sadoveanu, nr. 3, 700490–Iaşi, Romania Corresponding author: [email protected] This second part of the paper about the vascular flora of Rarău Massif listed approximately half of the whole number of the species registered by the authors in their field trips or already included in literature on the same area. Other taxa have been added to the initial list of plants, so that, the total number of taxa registered by the authors in Rarău Massif amount to 1443 taxa (1133 species and 310 subspecies, varieties and forms). There was signaled out the alien taxa on the surveyed area (18 species) and those dubious presence of some taxa for the same area (17 species). Also, there were listed all the vascular plants, protected by various laws or regulations, both internal or international, existing in Rarău (i.e. 189 taxa). Finally, there has been assessed the degree of wild flora conservation, using several indicators introduced in literature by Nowak, as they are: conservation indicator (C), threat conservation indicator) (CK), sozophytisation indicator (W), and conservation effectiveness indicator (E). Key words: Vascular flora, Rarău Massif, Romania, conservation indicators. 1. INTRODUCTION A comprehensive analysis of Rarău flora, in terms of plant diversity, taxonomic structure, biological, ecological and phytogeographic characteristics, as well as in terms of the richness in endemics, relict or threatened plant species was published in our previous note (see Oprea & Sîrbu 2012). -

Compositae Giseke (1792)
Multequina ISSN: 0327-9375 [email protected] Instituto Argentino de Investigaciones de las Zonas Áridas Argentina VITTO, LUIS A. DEL; PETENATTI, E. M. ASTERÁCEAS DE IMPORTANCIA ECONÓMICA Y AMBIENTAL. PRIMERA PARTE. SINOPSIS MORFOLÓGICA Y TAXONÓMICA, IMPORTANCIA ECOLÓGICA Y PLANTAS DE INTERÉS INDUSTRIAL Multequina, núm. 18, 2009, pp. 87-115 Instituto Argentino de Investigaciones de las Zonas Áridas Mendoza, Argentina Disponible en: http://www.redalyc.org/articulo.oa?id=42812317008 Cómo citar el artículo Número completo Sistema de Información Científica Más información del artículo Red de Revistas Científicas de América Latina, el Caribe, España y Portugal Página de la revista en redalyc.org Proyecto académico sin fines de lucro, desarrollado bajo la iniciativa de acceso abierto ISSN 0327-9375 ASTERÁCEAS DE IMPORTANCIA ECONÓMICA Y AMBIENTAL. PRIMERA PARTE. SINOPSIS MORFOLÓGICA Y TAXONÓMICA, IMPORTANCIA ECOLÓGICA Y PLANTAS DE INTERÉS INDUSTRIAL ASTERACEAE OF ECONOMIC AND ENVIRONMENTAL IMPORTANCE. FIRST PART. MORPHOLOGICAL AND TAXONOMIC SYNOPSIS, ENVIRONMENTAL IMPORTANCE AND PLANTS OF INDUSTRIAL VALUE LUIS A. DEL VITTO Y E. M. PETENATTI Herbario y Jardín Botánico UNSL, Cátedras Farmacobotánica y Famacognosia, Facultad de Química, Bioquímica y Farmacia, Universidad Nacional de San Luis, Ej. de los Andes 950, D5700HHW San Luis, Argentina. [email protected]. RESUMEN Las Asteráceas incluyen gran cantidad de especies útiles (medicinales, agrícolas, industriales, etc.). Algunas han sido domesticadas y cultivadas desde la Antigüedad y otras conforman vastas extensiones de vegetación natural, determinando la fisonomía de numerosos paisajes. Su uso etnobotánico ha ayudado a sustentar numerosos pueblos. Hoy, unos 40 géneros de Asteráceas son relevantes en alimentación humana y animal, fuentes de aceites fijos, aceites esenciales, forraje, miel y polen, edulcorantes, especias, colorantes, insecticidas, caucho, madera, leña o celulosa. -

Biogeographic History and Mating System of Rhodendron Ferrugineum in the French Pyrenees Olivia Charrier
Biogeographic history and mating system of Rhodendron ferrugineum in the French Pyrenees Olivia Charrier To cite this version: Olivia Charrier. Biogeographic history and mating system of Rhodendron ferrugineum in the French Pyrenees. Global Changes. INSA de Toulouse, 2014. English. NNT : 2014ISAT0034. tel-01204667 HAL Id: tel-01204667 https://tel.archives-ouvertes.fr/tel-01204667 Submitted on 24 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 5)µ4& &OWVFEFMPCUFOUJPOEV %0$503"5%&-6/*7&34*5²%&506-064& %ÏMJWSÏQBS Institut National des Sciences Appliquées de Toulouse (INSA de Toulouse) 1SÏTFOUÏFFUTPVUFOVFQBS Olivia Charrier -F 3 Octobre 2014 5Jtre : Histoire biogéographique et système de reproduction de Rhododendron ferrugineum dans les Pyrénées ED SEVAB : Écologie, biodiversité et évolution 6OJUÏEFSFDIFSDIF Laboratoire EDB (Evolution et Diversité Biologique) %JSFDUFVS T EFʾÒTF Dr Nathalie Escaravage et Dr André Pornon 3BQQPSUFVST M. Bertrand Schatz (DR CNRS Montpellier) Rapporteur M. Denis Michez (MCF UMH Mons - Belgique) Rapporteur "VUSF T NFNCSF T EVKVSZ Mme Myriam Gaudeul (MCF Museum Paris) Examinatrice Mme Sergine Ponsard (PR. UT3 Paul Sabatier) Présidente du jury Mme Nathalie Escaravage (MCF UT3 Paul Sabatier) Directrice de thèse REMERCIEMENTS -------------- Faire une thèse c’est un peu comme emprunter une route sinueuse de montagne, avec ses virages, ses cols à gravir et sa météo changeante.