Temporal Transcriptomic and Proteomic Landscapes of Deteriorating Pancreatic Islets in Type 2 Diabetic Rats
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Establishing the Pathogenicity of Novel Mitochondrial DNA Sequence Variations: a Cell and Molecular Biology Approach
Mafalda Rita Avó Bacalhau Establishing the Pathogenicity of Novel Mitochondrial DNA Sequence Variations: a Cell and Molecular Biology Approach Tese de doutoramento do Programa de Doutoramento em Ciências da Saúde, ramo de Ciências Biomédicas, orientada pela Professora Doutora Maria Manuela Monteiro Grazina e co-orientada pelo Professor Doutor Henrique Manuel Paixão dos Santos Girão e pela Professora Doutora Lee-Jun C. Wong e apresentada à Faculdade de Medicina da Universidade de Coimbra Julho 2017 Faculty of Medicine Establishing the pathogenicity of novel mitochondrial DNA sequence variations: a cell and molecular biology approach Mafalda Rita Avó Bacalhau Tese de doutoramento do programa em Ciências da Saúde, ramo de Ciências Biomédicas, realizada sob a orientação científica da Professora Doutora Maria Manuela Monteiro Grazina; e co-orientação do Professor Doutor Henrique Manuel Paixão dos Santos Girão e da Professora Doutora Lee-Jun C. Wong, apresentada à Faculdade de Medicina da Universidade de Coimbra. Julho, 2017 Copyright© Mafalda Bacalhau e Manuela Grazina, 2017 Esta cópia da tese é fornecida na condição de que quem a consulta reconhece que os direitos de autor são pertença do autor da tese e do orientador científico e que nenhuma citação ou informação obtida a partir dela pode ser publicada sem a referência apropriada e autorização. This copy of the thesis has been supplied on the condition that anyone who consults it recognizes that its copyright belongs to its author and scientific supervisor and that no quotation from the -

C6orf203 Controls OXPHOS Function Through Modulation of Mitochondrial Protein Biosynthesis
bioRxiv preprint doi: https://doi.org/10.1101/704403; this version posted July 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. C6orf203 controls OXPHOS function through modulation of mitochondrial protein biosynthesis number of characters excluding Materials and Methods: 40,651 Sara Palacios-Zambrano1,2, Luis Vázquez-Fonseca1,2, Cristina González-Páramos1,2, Laura Mamblona1,2, Laura Sánchez-Caballero3, Leo Nijtmans3, Rafael Garesse1,2 and Miguel Angel Fernández-Moreno1,2,* 1 Departamento de Bioquímica, Instituto de Investigaciones Biomédicas “Alberto Sols” UAM CSIC and Centro de Investigación Biomédica en Red en Enfermedades Raras (CIBERER). Facultad de Medicina, Universidad Autónoma de Madrid. Madrid 28029, Spain. 2 Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), Madrid 28041, Spain. 3 Department of Pediatrics, Radboud Center for Mitochondrial Medicine, Radboud University Medical Center, Nijmegen, The Netherlands. * To whom correspondence should be addressed. Tel:+34 91 497 31 29; Email: [email protected] Running title “C6orf203 controls mt-proteins synthesis” bioRxiv preprint doi: https://doi.org/10.1101/704403; this version posted July 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. ABSTRACT Mitochondria are essential organelles present in the vast majority of eukaryotic cells. Their central function is to produce cellular energy through the OXPHOS system, and functional alterations provoke so-called mitochondrial OXPHOS diseases. It is estimated that several hundred mitochondrial proteins have unknown functions. Very recently, C6orf203 was described to participate in mitochondrial transcription under induced mitochondrial DNA depletion stress conditions. -

A SARS-Cov-2 Protein Interaction Map Reveals Targets for Drug Repurposing
Article A SARS-CoV-2 protein interaction map reveals targets for drug repurposing https://doi.org/10.1038/s41586-020-2286-9 A list of authors and affiliations appears at the end of the paper Received: 23 March 2020 Accepted: 22 April 2020 A newly described coronavirus named severe acute respiratory syndrome Published online: 30 April 2020 coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19), has infected over 2.3 million people, led to the death of more than Check for updates 160,000 individuals and caused worldwide social and economic disruption1,2. There are no antiviral drugs with proven clinical efcacy for the treatment of COVID-19, nor are there any vaccines that prevent infection with SARS-CoV-2, and eforts to develop drugs and vaccines are hampered by the limited knowledge of the molecular details of how SARS-CoV-2 infects cells. Here we cloned, tagged and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identifed the human proteins that physically associated with each of the SARS-CoV-2 proteins using afnity-purifcation mass spectrometry, identifying 332 high-confdence protein–protein interactions between SARS-CoV-2 and human proteins. Among these, we identify 66 druggable human proteins or host factors targeted by 69 compounds (of which, 29 drugs are approved by the US Food and Drug Administration, 12 are in clinical trials and 28 are preclinical compounds). We screened a subset of these in multiple viral assays and found two sets of pharmacological agents that displayed antiviral activity: inhibitors of mRNA translation and predicted regulators of the sigma-1 and sigma-2 receptors. -

Transcriptomic and Proteomic Landscape of Mitochondrial
TOOLS AND RESOURCES Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals Inge Ku¨ hl1,2†*, Maria Miranda1†, Ilian Atanassov3, Irina Kuznetsova4,5, Yvonne Hinze3, Arnaud Mourier6, Aleksandra Filipovska4,5, Nils-Go¨ ran Larsson1,7* 1Department of Mitochondrial Biology, Max Planck Institute for Biology of Ageing, Cologne, Germany; 2Department of Cell Biology, Institute of Integrative Biology of the Cell (I2BC) UMR9198, CEA, CNRS, Univ. Paris-Sud, Universite´ Paris-Saclay, Gif- sur-Yvette, France; 3Proteomics Core Facility, Max Planck Institute for Biology of Ageing, Cologne, Germany; 4Harry Perkins Institute of Medical Research, The University of Western Australia, Nedlands, Australia; 5School of Molecular Sciences, The University of Western Australia, Crawley, Australia; 6The Centre National de la Recherche Scientifique, Institut de Biochimie et Ge´ne´tique Cellulaires, Universite´ de Bordeaux, Bordeaux, France; 7Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden Abstract Dysfunction of the oxidative phosphorylation (OXPHOS) system is a major cause of human disease and the cellular consequences are highly complex. Here, we present comparative *For correspondence: analyses of mitochondrial proteomes, cellular transcriptomes and targeted metabolomics of five [email protected] knockout mouse strains deficient in essential factors required for mitochondrial DNA gene (IKu¨ ); expression, leading to OXPHOS dysfunction. Moreover, -

Fatty Acid Metabolism Driven Mitochondrial Bioenergetics Promotes Advanced Developmental Phenotypes in Human Induced Pluripotent Stem Cell Derived Cardiomyocytes
Fatty acid metabolism driven mitochondrial bioenergetics promotes advanced developmental phenotypes in human induced pluripotent stem cell derived cardiomyocytes Chrishan J.A. Ramachandra1,a,b, Ashish Mehta1,c, Philip Wonga,b,d,e*, K.P. Myu Mai Jaa, Regina Fritsche-Danielsonf, Ratan V. Bhatg, Derek J. Hausenloya,b,h,i,j, Jean-Paul Kovalikb and Winston Shima,b,k* aNational Heart Research Institute Singapore, National Heart Centre Singapore bCardiovascular & Metabolic Disorders Program, Duke-NUS Medical School, Singapore cPSC and Phenotyping Laboratory, Victor Chang Cardiac Research Institute, Sydney, Australia dDepartment of Cardiology, National Heart Centre Singapore eSchool of Materials Science and Engineering, Nanyang Technological University, Singapore fCardiovascular and Metabolic Disease Innovative Medicines and Early Development Unit, AstraZeneca Research and Development, Gothenburg, Sweden gStrategy and External Innovation Department, AstraZeneca, Gothenburg, Sweden hThe Hatter Cardiovascular Institute, University College London, United Kingdom iBarts Heart Centre, St Barthlomew’s Hospital, London, United Kingdom jYong Loo Lin School of Medicine, National University of Singapore kHealth and Social Sciences Cluster, Singapore Institute of Technology 1Both authors contributed equally Running title: Cardiomyocyte metabolism and bioenergetics *Corresponding authors: Philip Wong National Heart Centre Singapore, 5 Hospital Drive, Singapore 169609 Email: [email protected]; Phone: +65 6704 8964; Fax: +65 6844 9053 Winston Shim -

Transcriptome Profiling Reveals the Complexity of Pirfenidone Effects in IPF
ERJ Express. Published on August 30, 2018 as doi: 10.1183/13993003.00564-2018 Early View Original article Transcriptome profiling reveals the complexity of pirfenidone effects in IPF Grazyna Kwapiszewska, Anna Gungl, Jochen Wilhelm, Leigh M. Marsh, Helene Thekkekara Puthenparampil, Katharina Sinn, Miroslava Didiasova, Walter Klepetko, Djuro Kosanovic, Ralph T. Schermuly, Lukasz Wujak, Benjamin Weiss, Liliana Schaefer, Marc Schneider, Michael Kreuter, Andrea Olschewski, Werner Seeger, Horst Olschewski, Malgorzata Wygrecka Please cite this article as: Kwapiszewska G, Gungl A, Wilhelm J, et al. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF. Eur Respir J 2018; in press (https://doi.org/10.1183/13993003.00564-2018). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2018 Copyright 2018 by the European Respiratory Society. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF Grazyna Kwapiszewska1,2, Anna Gungl2, Jochen Wilhelm3†, Leigh M. Marsh1, Helene Thekkekara Puthenparampil1, Katharina Sinn4, Miroslava Didiasova5, Walter Klepetko4, Djuro Kosanovic3, Ralph T. Schermuly3†, Lukasz Wujak5, Benjamin Weiss6, Liliana Schaefer7, Marc Schneider8†, Michael Kreuter8†, Andrea Olschewski1, -

Detection of H3k4me3 Identifies Neurohiv Signatures, Genomic
viruses Article Detection of H3K4me3 Identifies NeuroHIV Signatures, Genomic Effects of Methamphetamine and Addiction Pathways in Postmortem HIV+ Brain Specimens that Are Not Amenable to Transcriptome Analysis Liana Basova 1, Alexander Lindsey 1, Anne Marie McGovern 1, Ronald J. Ellis 2 and Maria Cecilia Garibaldi Marcondes 1,* 1 San Diego Biomedical Research Institute, San Diego, CA 92121, USA; [email protected] (L.B.); [email protected] (A.L.); [email protected] (A.M.M.) 2 Departments of Neurosciences and Psychiatry, University of California San Diego, San Diego, CA 92103, USA; [email protected] * Correspondence: [email protected] Abstract: Human postmortem specimens are extremely valuable resources for investigating trans- lational hypotheses. Tissue repositories collect clinically assessed specimens from people with and without HIV, including age, viral load, treatments, substance use patterns and cognitive functions. One challenge is the limited number of specimens suitable for transcriptional studies, mainly due to poor RNA quality resulting from long postmortem intervals. We hypothesized that epigenomic Citation: Basova, L.; Lindsey, A.; signatures would be more stable than RNA for assessing global changes associated with outcomes McGovern, A.M.; Ellis, R.J.; of interest. We found that H3K27Ac or RNA Polymerase (Pol) were not consistently detected by Marcondes, M.C.G. Detection of H3K4me3 Identifies NeuroHIV Chromatin Immunoprecipitation (ChIP), while the enhancer H3K4me3 histone modification was Signatures, Genomic Effects of abundant and stable up to the 72 h postmortem. We tested our ability to use H3K4me3 in human Methamphetamine and Addiction prefrontal cortex from HIV+ individuals meeting criteria for methamphetamine use disorder or not Pathways in Postmortem HIV+ Brain (Meth +/−) which exhibited poor RNA quality and were not suitable for transcriptional profiling. -

SUPPLEMENTARY MATERIALS and METHODS PBMC Transcriptomics
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY MATERIALS AND METHODS PBMC transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF Contents l Supplementary methods l Supplementary Figure 1 l Supplementary Figure 2 l Supplementary Figure 3 l Supplementary Figure 4 l Supplementary Figure 5 l Supplementary Table 1 l Supplementary Table 2 l Supplementary Table 3 l Supplementary Table 4 l Supplementary Tables 5-14 l Supplementary Table 15 l Supplementary Table 16 l Supplementary Table 17 Li J, et al. Gut 2021;0:1–13. doi: 10.1136/gutjnl-2020-323395 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY METHODS Test for HBV DNA The levels of HBV DNA were detected using real-time PCR with a COBAS® AmpliPrep/COBAS® TaqMan 48 System (Roche, Basel, Switzerland) and HBV Test v2.0. Criteria for diagnosing cirrhosis Pathology The gold standard for the diagnosis of cirrhosis is a liver biopsy obtained through a percutaneous or transjugular approach.1 Ultrasonography was performed 2-4 hours before biopsy. Liver biopsy specimens were obtained by experienced physicians. Percutaneous transthoracic puncture of the liver was performed according to the standard criteria. After biopsy, patients were monitored in the hospital with periodic analyses of haematocrit and other vital signs for 24 hours. Cirrhosis was diagnosed according to the globally agreed upon criteria.2 Cirrhosis is defined based on its pathological features under a microscope: (a) the presence of parenchymal nodules, (b) differences in liver cell size and appearance, (c) fragmentation of the biopsy specimen, (d) fibrous septa, and (d) an altered architecture and vascular relationships. -

UCLA UCLA Electronic Theses and Dissertations
UCLA UCLA Electronic Theses and Dissertations Title Matrix Processing Peptidase and Putative Roles in Mitochondrial Biogenesis. Nuc1 and Porin influence L-A Killer Virus Loads Co-dependently in Saccharomyces Cerevisiae. Permalink https://escholarship.org/uc/item/3600s4k4 Author Torres, Eric Publication Date 2019 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA Los Angeles Matrix Processing Peptidase and Putative Roles in Mitochondrial Biogenesis and Nuc1 and Porin influence L-A Killer Virus Loads Co-dependently in Saccharomyces Cerevisiae A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biochemistry and Molecular Biology by Eric Rommel Torres 2019 © Copyright by Eric Rommel Torres 2019 ABSTRACT OF THE DISSERTATION Matrix Processing Peptidase and Putative Roles in Mitochondrial Biogenesis and Nuc1 and Porin Influence Killer Virus Loads Co-dependently in Saccharomyces Cerevisiae by Eric Rommel Torres Doctor of Philosophy in Biochemistry and Molecular Biology University of California, Los Angeles, 2019 Professor Carla Marie Koehler, Chair Matrix Processing Peptidase and Putative Roles in Mitochondrial Biogenesis The mitochondrial protein import and proteolytic system are important for homeostasis of the mitochondrion. Typically, precursor proteins are targeted to the mitochondria by an N- terminal targeting sequence and are imported via TOM and TIM translocons of the outer and inner membrane. A series of proteases is required for the import and processing of precursors as well assembly and degradation of mitochondrial proteins. The matrix processing peptidase (MPP) coordinates the removal of presequences from precursors and is important in maintaining mitochondrial homeostasis. Furthermore, MPP is associated with neurodegenerative diseases. -

Mutations in the Substrate Binding Glycine-Rich Loop of the Mitochondrial Processing Peptidase-Α Protein (PMPCA) Cause a Severe Mitochondrial Disease
Mutations in the substrate binding glycine-rich loop of the mitochondrial processing peptidase-α protein (PMPCA) cause a severe mitochondrial disease The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Joshi, M., I. Anselm, J. Shi, T. A. Bale, M. Towne, K. Schmitz-Abe, L. Crowley, et al. 2016. “Mutations in the substrate binding glycine- rich loop of the mitochondrial processing peptidase-α protein (PMPCA) cause a severe mitochondrial disease.” Cold Spring Harbor Molecular Case Studies 2 (3): a000786. doi:10.1101/mcs.a000786. http://dx.doi.org/10.1101/mcs.a000786. Published Version doi:10.1101/mcs.a000786 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:27320259 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA COLD SPRING HARBOR Molecular Case Studies | RESEARCH ARTICLE Mutations in the substrate binding glycine-rich loop of the mitochondrial processing peptidase-α protein (PMPCA) cause a severe mitochondrial disease Mugdha Joshi,1,2,3,10 Irina Anselm,4,10 Jiahai Shi,5,6 Tejus A. Bale,7 Meghan Towne,1,3 Klaus Schmitz-Abe,1 Laura Crowley,1,3 Felix C. Giani,8,9 Shideh Kazerounian,1 Kyriacos Markianos,1 Hart G. Lidov,7 Rebecca Folkerth,7 Vijay G. Sankaran,8,9 and Pankaj B. Agrawal1,2,3 1Division of Genetics and Genomics, Boston Children’s -

The Endosome Is a Master Regulator of Plasma Membrane Collagen Fibril Assembly
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.25.436925; this version posted March 25, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The endosome is a master regulator of plasma membrane collagen fibril assembly 1Joan Chang*, 1Adam Pickard, 1Richa Garva, 1Yinhui Lu, 2Donald Gullberg and 1Karl E. Kadler* 1Wellcome Centre for Cell-Matrix Research, Faculty of Biology, Medical and Health, University of Manchester, Michael Smith Building, Oxford Road, Manchester M13 9PT UK, 2Department of Biomedicine and Center for Cancer Biomarkers, Norwegian Center of Excellence, University of Bergen, Norway. * Co-corresponding authors: JC email: [email protected] (orcid.org/0000-0002-7283- 9759); KEK email: [email protected] (orcid.org/0000-0003-4977-4683) Keywords: collagen-I, endocytosis, extracellular matrix, fibril, fibrillogenesis, integrin-a11, trafficking, VPS33b, [abstract] [149 word max] Collagen fibrils are the principal supporting elements in vertebrate tissues. They account for 25% of total protein mass, exhibit a broad range of size and organisation depending on tissue and stage of development, and can be under circadian clock control. Here we show that the remarkable dynamic pleomorphism of collagen fibrils is underpinned by a mechanism that distinguishes between collagen secretion and initiation of fibril assembly, at the plasma membrane. Collagen fibrillogenesis occurring at the plasma membrane requires vacuolar protein sorting (VPS) 33b (which is under circadian clock control), collagen-binding integrin-a11 subunit, and is reduced when endocytosis is inhibited. Fibroblasts lacking VPS33b secrete soluble collagen without assembling fibrils, whereas constitutive over-expression of VPS33b increases fibril number with loss of fibril rhythmicity. -
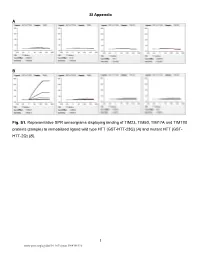
Sample) to Immobilized Ligand Wild Type HTT (GST-HTT-23Q) (A) and Mutant HTT (GST- HTT-2Q) (B)
SI Appendix A B Fig. S1. Representative SPR sensorgrams displaying binding of TIM23, TIM50, TIM17A and TIM17B proteins (sample) to immobilized ligand wild type HTT (GST-HTT-23Q) (A) and mutant HTT (GST- HTT-2Q) (B). 1 www.pnas.org/cgi/doi/10.1073/pnas.1904101116 Fig. S2. The level of mtCO1 protein is equal in equivalent number of mitochondria from ST-Hdh- Q7/Q7 and -Q111/Q111 cells. Representative immunoblot (A) and quantification (B) of mtCO1 level in FACS sorted mitochondria isolated from mito-eGFP transfected cells. Four million (4 MM) eGFP labeled mitochondria and 5 µg protein samples were immunoblotted for mtCO1, and nuclearly- encoded matrix aconitase 2 (ACO2) and outer membrane (SAM50) proteins. WCL – whole cell lysate, MITO – mitochondria, N.T. – not transfected, OMM – outer mitochondrial membrane. (n=3, data shown as mean+s.e.m. ** P<0.001, * P<0.05, t-Test). 2 A B 3 Fig. S3. Purity of mitochondria enriched fractions. (A) Representative immunoblot of Q7 and Q111 whole cell lysates (WCL), mitochondrial (Mito.) and cytosolic fractions (Cyto.) probed for mitochondrial markers (VDAC1, ATP5A, ACO2 and mtCO1) and typical mitochondrial fraction contaminants and contaminating organelles: cytosol (TUBA), endoplasmic reticulum (CALR), lysosomes (LAMP1), Golgi apparatus (RCAS1) and plasma membrane (CAV2). (B) Nonsynaptosomal (NS) and synaptosomal (SM) mitochondrial fractions isolated from fresh post- surgical cortex and frozen specimens of cortex and striatum (tissue blocks listed in Table 1) were tested for enrichment with mitochondrial markers and contamination with markers listed in (A) compared to whole brain homogenate (WB) and cytosolic fraction. 4 Fig. S4. Representative immunoblots of mitochondrial proteins from Q7 and Q111 cells (shown here to supplement Fig.