Unraveling the Specific Ischemic Core and Penumbra Transcriptome In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 As
cells Article Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 as New Binding Partners 1, 1, 2, Eunate Gallardo-Vara y, Lidia Ruiz-Llorente y, Juan Casado-Vela y , 3 4 5 6, , María J. Ruiz-Rodríguez , Natalia López-Andrés , Asit K. Pattnaik , Miguel Quintanilla z * 1, , and Carmelo Bernabeu z * 1 Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas (CSIC), and Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), 28040 Madrid, Spain; [email protected] (E.G.-V.); [email protected] (L.R.-L.) 2 Bioengineering and Aerospace Engineering Department, Universidad Carlos III and Centro de Investigación Biomédica en Red Enfermedades Neurodegenerativas (CIBERNED), Leganés, 28911 Madrid, Spain; [email protected] 3 Centro Nacional de Investigaciones Cardiovasculares (CNIC), 28029 Madrid, Spain; [email protected] 4 Cardiovascular Translational Research, Navarrabiomed, Complejo Hospitalario de Navarra (CHN), Universidad Pública de Navarra (UPNA), IdiSNA, 31008 Pamplona, Spain; [email protected] 5 School of Veterinary Medicine and Biomedical Sciences, and Nebraska Center for Virology, University of Nebraska-Lincoln, Lincoln, NE 68583, USA; [email protected] 6 Instituto de Investigaciones Biomédicas “Alberto Sols”, Consejo Superior de Investigaciones Científicas (CSIC), and Departamento de Bioquímica, Universidad Autónoma de Madrid (UAM), 28029 Madrid, Spain * Correspondence: [email protected] (M.Q.); [email protected] (C.B.) These authors contributed equally to this work. y Equal senior contribution. z Received: 7 August 2019; Accepted: 7 September 2019; Published: 13 September 2019 Abstract: Endoglin is a 180-kDa glycoprotein receptor primarily expressed by the vascular endothelium and involved in cardiovascular disease and cancer. -

SNP Analysis Reveals an Evolutionary Acceleration of the Human-Specific Micrornas
CORE Metadata, citation and similar papers at core.ac.uk Provided by Nature Precedings Rapid evolution of the human-specific microRNAs SNP analysis reveals an evolutionary acceleration of the human-specific microRNAs Qipeng Zhang1, 2, Ming Lu1, 2, and Qinghua Cui1, 2 1. Department of Medical Informatics, Peking University Health Science Center, Peking University, 38 Xueyuan Rd, Beijing, China 100083 2. Ministry of Education Key Lab of Molecular Cardiovascular Sciences, Peking University, 38 Xueyuan Rd, Beijing, China 100083 Nature Precedings : hdl:10101/npre.2008.2127.1 Posted 29 Jul 2008 Corresponding author: Cui, Q. ([email protected]) 1 PDF 文件使用 "pdfFactory Pro" 试用版本创建 www.fineprint.cn Rapid evolution of the human-specific microRNAs MicroRNAs are one class of important gene regulators at the post-transcriptional level by binding to the 3’UTRs of target mRNAs. It has been reported that human microRNAs are evolutionary conserved and show lower single nucleotide polymorphisms (SNPs) than their flanking regions. However, in this study, we report that the human-specific microRNAs show a higher SNP density than both the conserved microRNAs and other control regions, suggesting rapid evolution and positive selection has occurred in these regions. Furthermore, we observe that the human-specific microRNAs show greater SNPs minor allele frequency and the SNPs in the human-specific microRNAs show fewer effects on the stability of the microRNA secondary structure, indicating that the SNPs in the human-specific microRNAs tend to be less deleterious. Finally, two microRNAs hsa-mir-423 (SNP: rs6505162), hsa-mir-608 (SNP: rs4919510) and 288 target genes that have apparently been under recent positive selection are identified. -

WO 2017/214553 Al 14 December 2017 (14.12.2017) W !P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2017/214553 Al 14 December 2017 (14.12.2017) W !P O PCT (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, C12N 15/11 (2006.01) C12N 15/113 (2010.01) CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, (21) International Application Number: HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, PCT/US20 17/036829 KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (22) International Filing Date: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 09 June 2017 (09.06.2017) OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY,TH, TJ, TM, TN, (25) Filing Language: English TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (26) Publication Language: English (84) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of regional protection available): ARIPO (BW, GH, 62/347,737 09 June 2016 (09.06.2016) US GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, 62/408,639 14 October 2016 (14.10.2016) US UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/433,770 13 December 2016 (13.12.2016) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, (71) Applicant: THE GENERAL HOSPITAL CORPO¬ MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, RATION [US/US]; 55 Fruit Street, Boston, Massachusetts TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, 021 14 (US). -

The Disruption Ofcelf6, a Gene Identified by Translational Profiling
2732 • The Journal of Neuroscience, February 13, 2013 • 33(7):2732–2753 Neurobiology of Disease The Disruption of Celf6, a Gene Identified by Translational Profiling of Serotonergic Neurons, Results in Autism-Related Behaviors Joseph D. Dougherty,1,2 Susan E. Maloney,1,2 David F. Wozniak,2 Michael A. Rieger,1,2 Lisa Sonnenblick,3,4,5 Giovanni Coppola,4 Nathaniel G. Mahieu,1 Juliet Zhang,6 Jinlu Cai,8 Gary J. Patti,1 Brett S. Abrahams,8 Daniel H. Geschwind,3,4,5 and Nathaniel Heintz6,7 Departments of 1Genetics and 2Psychiatry, Washington University School of Medicine, St. Louis, Missouri 63110, 3UCLA Center for Autism Research and Treatment, Semel Institute for Neuroscience and Behavior, 4Program in Neurogenetics, Department of Neurology, and 5Department of Human Genetics, David Geffen School of Medicine at UCLA, Los Angeles, California 90095, 6Laboratory of Molecular Biology, Howard Hughes Medical Institute, and 7The GENSAT Project, Rockefeller University, New York, New York 10065, and 8Departments of Genetics and Neuroscience, Albert Einstein College of Medicine, New York, New York 10461 The immense molecular diversity of neurons challenges our ability to understand the genetic and cellular etiology of neuropsychiatric disorders. Leveraging knowledge from neurobiology may help parse the genetic complexity: identifying genes important for a circuit that mediates a particular symptom of a disease may help identify polymorphisms that contribute to risk for the disease as a whole. The serotonergic system has long been suspected in disorders that have symptoms of repetitive behaviors and resistance to change, including autism. We generated a bacTRAP mouse line to permit translational profiling of serotonergic neurons. -

Physiology and Molecular Characterization of Metabolism Related Mouse Models for Bone Disease
TECHNISCHE UNIVERSITÄT MÜNCHEN Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt Lehrstuhl für Experimentelle Genetik Physiology and molecular characterization of metabolism related mouse models for bone disease Shen Chi Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ.-Prof. Dr. W. Wurst Prüfer der Dissertation: 1. Univ.-Prof. Dr. M. Hrabé de Angelis 2. apl. Prof. Dr. J. Adamski Die Dissertation wurde am 16.03.2015 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 29.07.2015 angenommen. I. Table of contents I. TABLE OF CONTENTS .............................................................................................................. I II. FIGURES AND TABLES ........................................................................................................... IV III. ABBREVIATIONS ................................................................................................................ VII IV. ACKNOWLEDGEMENTS .................................................................................................. XII V. AFFIRMATION ......................................................................................................................... XII 1. SUMMARY/ZUSAMMENFASSUNG -

Supporting Information
Supporting Information Friedman et al. 10.1073/pnas.0812446106 SI Results and Discussion intronic miR genes in these protein-coding genes. Because in General Phenotype of Dicer-PCKO Mice. Dicer-PCKO mice had many many cases the exact borders of the protein-coding genes are defects in additional to inner ear defects. Many of them died unknown, we searched for miR genes up to 10 kb from the around birth, and although they were born at a similar size to hosting-gene ends. Out of the 488 mouse miR genes included in their littermate heterozygote siblings, after a few weeks the miRBase release 12.0, 192 mouse miR genes were found as surviving mutants were smaller than their heterozygote siblings located inside (distance 0) or in the vicinity of the protein-coding (see Fig. 1A) and exhibited typical defects, which enabled their genes that are expressed in the P2 cochlear and vestibular SE identification even before genotyping, including typical alopecia (Table S2). Some coding genes include huge clusters of miRNAs (in particular on the nape of the neck), partially closed eyelids (e.g., Sfmbt2). Other genes listed in Table S2 as coding genes are [supporting information (SI) Fig. S1 A and C], eye defects, and actually predicted, as their transcript was detected in cells, but weakness of the rear legs that were twisted backwards (data not the predicted encoded protein has not been identified yet, and shown). However, while all of the mutant mice tested exhibited some of them may be noncoding RNAs. Only a single protein- similar deafness and stereocilia malformation in inner ear HCs, coding gene that is differentially expressed in the cochlear and other defects were variable in their severity. -

Epigenome-450K-Wide Methylation Signatures of Active Cigarette Smoking: the Young Finns Study
Bioscience Reports (2020) 40 BSR20200596 https://doi.org/10.1042/BSR20200596 Research Article Epigenome-450K-wide methylation signatures of active cigarette smoking: The Young Finns Study Pashupati P. Mishra1,2,3,*, Ismo Hanninen¨ 1,2,3,*, Emma Raitoharju1,2,3, Saara Marttila1,2,3,4, Binisha H. Mishra1,2,3, Nina Mononen1,2,3,MikaKah¨ onen¨ 2,5, Mikko Hurme4,6, Olli Raitakari7,8,9, Petri Tor¨ onen¨ 10, Liisa Holm10,11 and Terho Lehtimaki¨ 1,2,3 Downloaded from http://portlandpress.com/bioscirep/article-pdf/40/7/BSR20200596/887717/bsr-2020-0596.pdf by guest on 27 September 2021 1Department of Clinical Chemistry, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland; 2Finnish Cardiovascular Research Center-Tampere, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland; 3Department of Clinical Chemistry, Fimlab Laboratories, Tampere, Finland; 4Gerontology Research Center (GEREC), Tampere University, Tampere, Finland; 5Department of Clinical Physiology, Tampere University Hospital, Tampere, Finland; 6Department of Microbiology and Immunology, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland; 7Centre for Population Health Research, University of Turku and Turku University Hospital, Turku, Finland; 8Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland; 9Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland; 10Institute of Biotechnology, Helsinki Institute of Life Sciences (HiLife), University of Helsinki, Helsinki, Finland; 11Organismal and Evolutionary Biology Research Program, Faculty of Biological and Environmental Sciences, University of Helsinki, Helsinki, Finland Correspondence: Pashupati P. Mishra (pashupati.mishra@tuni.fi) Smoking as a major risk factor for morbidity affects numerous regulatory systems of the human body including DNA methylation. -

Mrna Expression in Human Leiomyoma and Eker Rats As Measured by Microarray Analysis
Table 3S: mRNA Expression in Human Leiomyoma and Eker Rats as Measured by Microarray Analysis Human_avg Rat_avg_ PENG_ Entrez. Human_ log2_ log2_ RAPAMYCIN Gene.Symbol Gene.ID Gene Description avg_tstat Human_FDR foldChange Rat_avg_tstat Rat_FDR foldChange _DN A1BG 1 alpha-1-B glycoprotein 4.982 9.52E-05 0.68 -0.8346 0.4639 -0.38 A1CF 29974 APOBEC1 complementation factor -0.08024 0.9541 -0.02 0.9141 0.421 0.10 A2BP1 54715 ataxin 2-binding protein 1 2.811 0.01093 0.65 0.07114 0.954 -0.01 A2LD1 87769 AIG2-like domain 1 -0.3033 0.8056 -0.09 -3.365 0.005704 -0.42 A2M 2 alpha-2-macroglobulin -0.8113 0.4691 -0.03 6.02 0 1.75 A4GALT 53947 alpha 1,4-galactosyltransferase 0.4383 0.7128 0.11 6.304 0 2.30 AACS 65985 acetoacetyl-CoA synthetase 0.3595 0.7664 0.03 3.534 0.00388 0.38 AADAC 13 arylacetamide deacetylase (esterase) 0.569 0.6216 0.16 0.005588 0.9968 0.00 AADAT 51166 aminoadipate aminotransferase -0.9577 0.3876 -0.11 0.8123 0.4752 0.24 AAK1 22848 AP2 associated kinase 1 -1.261 0.2505 -0.25 0.8232 0.4689 0.12 AAMP 14 angio-associated, migratory cell protein 0.873 0.4351 0.07 1.656 0.1476 0.06 AANAT 15 arylalkylamine N-acetyltransferase -0.3998 0.7394 -0.08 0.8486 0.456 0.18 AARS 16 alanyl-tRNA synthetase 5.517 0 0.34 8.616 0 0.69 AARS2 57505 alanyl-tRNA synthetase 2, mitochondrial (putative) 1.701 0.1158 0.35 0.5011 0.6622 0.07 AARSD1 80755 alanyl-tRNA synthetase domain containing 1 4.403 9.52E-05 0.52 1.279 0.2609 0.13 AASDH 132949 aminoadipate-semialdehyde dehydrogenase -0.8921 0.4247 -0.12 -2.564 0.02993 -0.32 AASDHPPT 60496 aminoadipate-semialdehyde -

STAT3 Targets Suggest Mechanisms of Aggressive Tumorigenesis in Diffuse Large B Cell Lymphoma
STAT3 Targets Suggest Mechanisms of Aggressive Tumorigenesis in Diffuse Large B Cell Lymphoma Jennifer Hardee*,§, Zhengqing Ouyang*,1,2,3, Yuping Zhang*,4 , Anshul Kundaje*,†, Philippe Lacroute*, Michael Snyder*,5 *Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305; §Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06520; and †Department of Computer Science, Stanford University School of Engineering, Stanford, CA 94305 1The Jackson Laboratory for Genomic Medicine, Farmington, CT 06030 2Department of Biomedical Engineering, University of Connecticut, Storrs, CT 06269 3Department of Genetics and Developmental Biology, University of Connecticut Health Center, Farmington, CT 06030 4Department of Biostatistics, Yale School of Public Health, Yale University, New Haven, CT 06520 5Corresponding author: Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305. Email: [email protected] DOI: 10.1534/g3.113.007674 Figure S1 STAT3 immunoblotting and immunoprecipitation with sc-482. Western blot and IPs show a band consistent with expected size (88 kDa) of STAT3. (A) Western blot using antibody sc-482 versus nuclear lysates. Lanes contain (from left to right) lysate from K562 cells, GM12878 cells, HeLa S3 cells, and HepG2 cells. (B) IP of STAT3 using sc-482 in HeLa S3 cells. Lane 1: input nuclear lysate; lane 2: unbound material from IP with sc-482; lane 3: material IP’d with sc-482; lane 4: material IP’d using control rabbit IgG. Arrow indicates the band of interest. (C) IP of STAT3 using sc-482 in K562 cells. Lane 1: input nuclear lysate; lane 2: material IP’d using control rabbit IgG; lane 3: material IP’d with sc-482. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses by a Synthetic TLR3 Ligand Mitigates
Downloaded from http://www.jimmunol.org/ by guest on September 28, 2021 Inducing is online at: average * The Journal of Immunology published online 16 August 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/08/16/jimmun ol.1300376 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 28, 2021. Published August 16, 2013, doi:10.4049/jimmunol.1300376 The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -
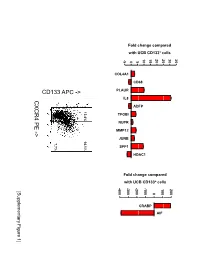
Supplementary Data
Fold change compared with UCB CD133+ cells 10 15 20 25 30 35 -5 0 5 COL4A1 CD68 PLAUR IL8 CXCR4 PE ADFP 13.4% 14.1% TFGBI PE NUPR - MMP12 > JUNB 7.7% SPP1 HDAC1 Fold change compared with UCB CD133+ cells -400 -300 -200 -100 100 200 [Supplementary Figure1] 0 CRABP AIF Rutella S et al. Supplementary Materials and Methods Real-time PCR Real-time quantitative PCR (qPCR) was performed on mRNA samples isolated from either endometrial cancer-derived CD133+ cells or umbilical cord blood CD133+ cells with the RNeasy plus Mini Kit (QIAGEN, Hilden, Germany), as already detailed. Quantitative PCR for a selected group of genes was performed on the iCycler iQ system (Bio-Rad, Hercules, CA). Primer sets were designed using the Beacon Design Software (Version 3) and the sequences available in the GeneBankTM data base. The specific oligonucleotide primer sequences are detailed in Supplementary Table 8. Complementary DNA (cDNA) was prepared starting from 1μg of total RNA using the iScrypt cDNA Synthesis Kit, which contains RNase H + MMLV reverse transcriptase random primers and 5x reaction mixture (Bio-Rad), according to the manufacturer’s instructions. Reactions were conducted in the PTC-0200 DNA Engine (MJ RESEARCH). Amplification was carried out in a total volume of 25μl containing 0.3μM of each specific primer, 12,5μl 2X SYBR Green Master Mix (100mM KCl, 40mM Tris-HCl, pH 8.4, 0.4mM of each dNTP, iTaq DNA polymerase, 6mM MgCl2, SYBR Green I, 20nM fluorescein, stabilizers; Bio-Rad) and 2μl of diluted cDNA. The PCR reactions were cycled starting with a 3-minute template denaturation step at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. -

Using Genetic Data in the Add Health Sample Andrew Smolen and Brett Haberstick Institute for Behavioral Genetics, University of Colorado at Boulder
Using genetic data in the Add Health Sample Andrew Smolen and Brett Haberstick Institute for Behavioral Genetics, University of Colorado at Boulder Add Health Users Workshop, July 2010 Bethesda, Maryland VNTR polymorphism of Dopamine Transporter 40 bp VNTR 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 3’ - UTR 5’ AAA 3’ Variable number of tandem repeats 3-13 copies Adapted from Miller & Madras, 2002; Fuke et al., (2001), Vandenberg et al., 1992 DRD4 VNTR Polymorphism in Exon 3 48 bp VNTR Translation start site 5’ +1 3’ EX1 EX2 EX3 EX4 EX5 Exon 3 VNTR 5’ AAA 3’ Variable number of tandem repeats 2-11 copies Adapted from: D’Souza; van Tol et al., (1992) Serotonin Transporter, SLC6A4 17q11.1-q12 minus strand [25,392,812 [25,685,191 EFCAB5 SLC6A4 TMIGD1 CCDC55 BLMH rs2066713 25,586,831] 25,549,032] 5’ 3’ 5HTTLPR 1a 1b 2 3 4 5 6 7 8 9 10 11 12 13 14 rs6354 rs1042173 rs12945042 rs140700 rs2054847 rs2020942 rs25531 MspI LA/LG Coding Region Untranslated Region VNTR and Ins/del Genotyping on ABI 3130xl DAT1-9R DRD4-4R Amelogenin MAOA-u DAT1-10R (male) 5HTTLPR-L DRD4-7R Amelogenin DRD4-4R (male) 5HTTLPR-S MAOA-u 5HTTLPR-L DAT1-10R 5HTTLPR-L Amelogenin MAOA-u DRD4-4R DAT1-10R (female) MAOA-u DRD4-4R 5HTTLPR-L Amelogenin 5HTTLPR-S DAT1-9R (female) DAT1-10R Genotyping four typical VNTR candidate genes Dinucleotide Repeat Genotyping on ABI 3130xl DRD5 MAOA STR 5’UTR STR Amelogenin intron 2 female MAOA STR DRD5 Amelogenin intron 2 5’UTR STR male MAOA STR DRD5 Amelogenin 5’UTR STR female intron 2 DRD4 Exon-3 48 bp VNTR 3R 2R 427 bp 379 bp 4R 475 bp 6R 571 bp 5R