Divergent Regulation of Myotube Formation and Gene Expression by E2 and EPA During In-Vitro Differentiation of C2C12 Myoblasts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Alveolar Rhabdomyosarcoma-Associated Proteins PAX3/FOXO1A and PAX7/FOXO1A Suppress the Transcriptional Activity of Myod-Target Genes in Muscle Stem Cells
Oncogene (2013) 32, 651 --662 & 2013 Macmillan Publishers Limited All rights reserved 0950-9232/13 www.nature.com/onc ORIGINAL ARTICLE Alveolar rhabdomyosarcoma-associated proteins PAX3/FOXO1A and PAX7/FOXO1A suppress the transcriptional activity of MyoD-target genes in muscle stem cells F Calhabeu1, S Hayashi2, JE Morgan3, F Relaix2 and PS Zammit1 Rhabdomyosarcoma (RMS) is the commonest soft-tissue sarcoma in childhood and is characterized by expression of myogenic proteins, including the transcription factors MyoD and myogenin. There are two main subgroups, embryonal RMS and alveolar RMS (ARMS). Most ARMS are associated with chromosomal translocations that have breakpoints in introns of either PAX3 or PAX7, and FOXO1A. These translocations create chimeric transcription factors termed PAX3/FOXO1A and PAX7/FOXO1A respectively. Upon ectopic PAX3/FOXO1A expression, together with other genetic manipulation in mice, both differentiating myoblasts and satellite cells (the resident stem cells of postnatal muscle) can give rise to tumours with ARMS characteristics. As PAX3 and PAX7 are part of transcriptional networks that regulate muscle stem cell function in utero and during early postnatal life, PAX3/FOXO1A and PAX7/FOXO1A may subvert normal PAX3 and PAX7 functions. Here we examined how PAX3/FOXO1A and PAX7/FOXO1A affect myogenesis in satellite cells. PAX3/FOXO1A or PAX7/FOXO1A inhibited myogenin expression and prevented terminal differentiation in murine satellite cells: the same effect as dominant-negative (DN) Pax3 or Pax7 constructs. The transcription of MyoD-target genes myogenin and muscle creatine kinase were suppressed by PAX3/FOXO1A or PAX7/ FOXO1A in C2C12 myogenic cells again as seen with Pax3/7DN. PAX3/FOXO1A or PAX7/FOXO1A did not inhibit the transcriptional activity of MyoD by perturbing MyoD expression, localization, phosphorylation or interaction with E-proteins. -

Microarray-Based Analysis of Genes, Transcription Factors, and Epigenetic Modifications in Lung Cancer Exposed to Nitric Oxide
CANCER GENOMICS & PROTEOMICS 17 : 401-415 (2020) doi:10.21873/cgp.20199 Microarray-based Analysis of Genes, Transcription Factors, and Epigenetic Modifications in Lung Cancer Exposed to Nitric Oxide ARNATCHAI MAIUTHED 1, ORNJIRA PRAKHONGCHEEP 2,3 and PITHI CHANVORACHOTE 2,3 1Department of Pharmacology, Faculty of Pharmacy, Mahidol University, Bangkok, Thailand; 2Cell-based Drug and Health Product Development Research Unit, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok, Thailand; 3Department of Pharmacology and Physiology, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok, Thailand Abstract. Background/Aim: Nitric oxide (NO) is recognized NO exposure of lung cancer cells resulted in a change in as an important biological mediator that exerts several transcription factors (TFs) and epigenetic modifications human physiological functions. As its nature is an aqueous (histone modification and miRNA). Interestingly, NO soluble gas that can diffuse through cells and tissues, NO treatment was shown to potentiate cancer stem cell-related can affect cell signaling, the phenotype of cancer and modify genes and transcription factors Oct4, Klf4, and Myc. surrounding cells. The variety of effects of NO on cancer cell Conclusion: Through this comprehensive approach, the biology has convinced researchers to determine the defined present study illustrated the scheme of how NO affects mechanisms of these effects and how to control this mediator molecular events in lung cancer cells. for a better understanding as well as for therapeutic gain. Materials and Methods: We used bioinformatics and Lung cancer is one of the leading causes of cancer-related pharmacological experiments to elucidate the potential deaths worldwide (1, 2). Since the aggressive phenotypes regulation and underlying mechanisms of NO in non-small and treatment failures have been frequently observed in lung a lung cancer cell model. -

Pig Antibodies
Pig Antibodies gene_name sku Entry_Name Protein_Names Organism Length Identity CDX‐2 ARP31476_P050 D0V4H7_PIG Caudal type homeobox 2 (Fragment) Sus scrofa (Pig) 147 100.00% CDX‐2 ARP31476_P050 A7MAE3_PIG Caudal type homeobox transcription factor 2 (Fragment) Sus scrofa (Pig) 75 100.00% Tnnt3 ARP51286_P050 Q75NH3_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 271 85.00% Tnnt3 ARP51286_P050 Q75NH2_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 266 85.00% Tnnt3 ARP51286_P050 Q75NH1_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 260 85.00% Tnnt3 ARP51286_P050 Q75NH0_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 250 85.00% Tnnt3 ARP51286_P050 Q75NG8_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 266 85.00% Tnnt3 ARP51286_P050 Q75NG7_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 260 85.00% Tnnt3 ARP51286_P050 Q75NG6_PIG Troponin T fast skeletal muscle type Sus scrofa (Pig) 250 85.00% Tnnt3 ARP51286_P050 TNNT3_PIG Troponin T, fast skeletal muscle (TnTf) Sus scrofa (Pig) 271 85.00% ORF Names:PANDA_000462 EMBL EFB13877.1OrganismAiluropod High mobility group protein B2 (High mobility group protein a melanoleuca (Giant panda) ARP31939_P050 HMGB2_PIG 2) (HMG‐2) Sus scrofa (Pig) 210 100.00% Agpat5 ARP47429_P050 B8XTR3_PIG 1‐acylglycerol‐3‐phosphate O‐acyltransferase 5 Sus scrofa (Pig) 365 85.00% irf9 ARP31200_P050 Q29390_PIG Transcriptional regulator ISGF3 gamma subunit (Fragment) Sus scrofa (Pig) 57 100.00% irf9 ARP31200_P050 Q0GFA1_PIG Interferon regulatory factor 9 Sus scrofa (Pig) -

Transcriptional Code and Disease Map for Adult Retinal Cell Types
RE so UR C E Transcriptional code and disease map for adult retinal cell types Sandra Siegert1,7, Erik Cabuy1,7, Brigitte Gross Scherf1, Hubertus Kohler1, Satchidananda Panda2, Yun-Zheng Le3,4, Hans Jörg Fehling5, Dimos Gaidatzis1,6, Michael B Stadler1,6 & Botond Roska1 Brain circuits are assembled from a large variety of morphologically and functionally diverse cell types. It is not known how the intermingled cell types of an individual adult brain region differ in their expressed genomes. Here we describe an atlas of cell type transcriptomes in one brain region, the mouse retina. We found that each adult cell type expressed a specific set of genes, including a unique set of transcription factors, forming a ‘barcode’ for cell identity. Cell type transcriptomes carried enough information to categorize cells into morphological classes and types. Several genes that were specifically expressed in particular retinal circuit elements, such as inhibitory neuron types, are associated with eye diseases. The resource described here allows gene expression to be compared across adult retinal cell types, experimenting with specific transcription factors to differentiate stem or somatic cells to retinal cell types, and predicting cellular targets of newly discovered disease-associated genes. The brain is composed of many neuronal cell types that are deter- across the retina with a mosaic-like distribution (Supplementary Text mined during development by a dynamic transcriptional program1–5. and Supplementary Fig. 1). The cellular architecture of the retina is In adults, neurons sampled from different brain areas such as the highly conserved among mammals14–17. cortex, cerebellum and hippocampus maintain differences in their By constructing and analyzing a transcriptome atlas for retinal cell expressed genomes6,7. -

Engineered Type 1 Regulatory T Cells Designed for Clinical Use Kill Primary
ARTICLE Acute Myeloid Leukemia Engineered type 1 regulatory T cells designed Ferrata Storti Foundation for clinical use kill primary pediatric acute myeloid leukemia cells Brandon Cieniewicz,1* Molly Javier Uyeda,1,2* Ping (Pauline) Chen,1 Ece Canan Sayitoglu,1 Jeffrey Mao-Hwa Liu,1 Grazia Andolfi,3 Katharine Greenthal,1 Alice Bertaina,1,4 Silvia Gregori,3 Rosa Bacchetta,1,4 Norman James Lacayo,1 Alma-Martina Cepika1,4# and Maria Grazia Roncarolo1,2,4# Haematologica 2021 Volume 106(10):2588-2597 1Department of Pediatrics, Division of Stem Cell Transplantation and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 2Stanford Institute for Stem Cell Biology and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 3San Raffaele Telethon Institute for Gene Therapy, Milan, Italy and 4Center for Definitive and Curative Medicine, Stanford School of Medicine, Stanford, CA, USA *BC and MJU contributed equally as co-first authors #AMC and MGR contributed equally as co-senior authors ABSTRACT ype 1 regulatory (Tr1) T cells induced by enforced expression of interleukin-10 (LV-10) are being developed as a novel treatment for Tchemotherapy-resistant myeloid leukemias. In vivo, LV-10 cells do not cause graft-versus-host disease while mediating graft-versus-leukemia effect against adult acute myeloid leukemia (AML). Since pediatric AML (pAML) and adult AML are different on a genetic and epigenetic level, we investigate herein whether LV-10 cells also efficiently kill pAML cells. We show that the majority of primary pAML are killed by LV-10 cells, with different levels of sensitivity to killing. Transcriptionally, pAML sensitive to LV-10 killing expressed a myeloid maturation signature. -
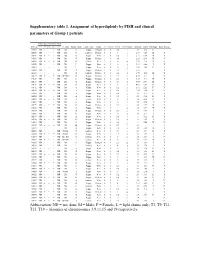
Supplementary Table 1. Assignment of Hyperdiploidy by FISH and Clinical Parameters of Group 1 Patients
Supplementary table 1. Assignment of hyperdiploidy by FISH and clinical parameters of Group 1 patients FISH Based Trisomy Index MAYO T3 T9 T11 T15 T19 TC class Heavy Chain Light Chain Status 13 VH/CH PCLI GEP Cluster Albumin B2M ISS Stage Bone Disease M003 ND + + + ND D1 G Kappa Relapse 0 0 4.2 1 3.4 4.16 II - M029 ND + - + ND D1 G lambda Relapse 0 0 3 1 2.74 7.29 III Y M037 ND + + - ND D2 G Kappa New 1 0 0.2 2 2.77 6.54 III Y M051 ND + - + ND D2 G Kappa Relapse 1 0 4.8 1 3.24 8.46 III Y M058 ND + + + ND D1 G lambda New 1 0 0 4 3.15 7.18 III N M059 ND + - + ND D1 L Kappa New 0 1 0 4 3.94 2.28 I Y M061 + + - - - D1 L Kappa New 0 0 0 2 2.67 2.61 II Y M082 ND + + - ND D2 A Kappa Relapse 1 0 5.8 1 2.76 - - Y M113 + + - - - D1 G lambda Relapse 0 0 2.2 4 3.79 11.6 III Y M123 ND + - + ND D1+D2 G Kappa Relapse 1 1 1.2 2 4.28 1.7 I Y M128 ND + + + ND D1 L Kappa Relapse 0 1 2.3 4 4.17 7.4 III Y M131 ND - + + ND D1 G Kappa Relapse 0 0 4.4 4 4.88 8.74 III Y M135 ND + + + ND D1 G Kappa New 0 0 0 3 4.01 5.23 II Y M142 ND + + - ND D1 A Kappa New 0 0 0.2 3 3.92 5.23 II Y M163 ND + + + ND D1 A Kappa New 0 0 0.4 3 2.96 2.06 II N M198 ND + + + ND D1 A Kappa New 0 0 0.4 2 3.1 3.3 II Y M200 ND + + - ND D2 A Kappa New 0 0 3.8 1 2.5 3.16 II Y M218 ND + + + ND D1 A lambda New 0 0 0 3 3.6 4.31 II Y M227 ND - + + ND D1 L Kappa New 1 0 0 3 3.9 2.76 I Y M229 ND - + + ND D1 G Kappa Relapse 0 0 3.8 1 4.3 8.77 III Y M230 ND + + - ND D1 A Kappa New 1 1 1 4 3.6 6.94 III Y M235 ND + + + ND D1 G Kappa Relapse 0 0 0.2 2 4 2.37 I Y M245 ND + - + ND D1 G Kappa New 0 0 0 4 -

Discovery of Biased Orientation of Human DNA Motif Sequences
bioRxiv preprint doi: https://doi.org/10.1101/290825; this version posted January 27, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Discovery of biased orientation of human DNA motif sequences 2 affecting enhancer-promoter interactions and transcription of genes 3 4 Naoki Osato1* 5 6 1Department of Bioinformatic Engineering, Graduate School of Information Science 7 and Technology, Osaka University, Osaka 565-0871, Japan 8 *Corresponding author 9 E-mail address: [email protected], [email protected] 10 1 bioRxiv preprint doi: https://doi.org/10.1101/290825; this version posted January 27, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 11 Abstract 12 Chromatin interactions have important roles for enhancer-promoter interactions 13 (EPI) and regulating the transcription of genes. CTCF and cohesin proteins are located 14 at the anchors of chromatin interactions, forming their loop structures. CTCF has 15 insulator function limiting the activity of enhancers into the loops. DNA binding 16 sequences of CTCF indicate their orientation bias at chromatin interaction anchors – 17 forward-reverse (FR) orientation is frequently observed. DNA binding sequences of 18 CTCF were found in open chromatin regions at about 40% - 80% of chromatin 19 interaction anchors in Hi-C and in situ Hi-C experimental data. -

Characterizing Thyroid Hormone Mediated Action on Gene
Characterizing thyroid hormone mediated action on gene expression in mice: mechanistic insight into thyroid hormone response elements, thyroid hormone receptor-binding sites, and microRNAs by Martin A. Paquette A thesis submitted to the Faculty of Graduate and Postdoctoral Affairs in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biology Carleton University Ottawa, Ontario ©2013 Martin A. Paquette ABSTRACT Thyroid hormone (TH) exerts its effects by binding to the TH receptor (TR), which binds to TH response elements (TREs) to regulate target gene expression. Disruption of TH action can have detrimental health effects. The precise molecular mechanisms involved in TH mediated gene expression remain unclear. The overall objectives of this thesis were to: i) characterize global gene and microRNA (miRNA) expression in early response to TH perturbation in mouse liver; ii) identify TREs and TR-binding sites found throughout the mouse genome; and iii) compare TRE half-site organizations and their ability to drive gene expression. Transcriptional profiling of mRNA liver samples from TH disrupted mice enabled the identification of genes that were under direct TH-regulation. TREs in the promoter region of Tor1a, Hectd3, Slc25a45 and 2310003H01Rik were validated in vitro, adding four genes to the battery of only 13 known TRE- containing mouse genes. Hepatic miRNAs were also found to be significantly altered following perturbations in TH levels. In vitro analyses confirmed TH regulation of miR-206. Moreover, Mup1 and Gpd2 were confirmed to be targeted by miR-206 in response to TH, demonstrating that miRNAs can act as master regulators of the TH response pathway. -

Table S3. RAE Analysis of Well-Differentiated Liposarcoma
Table S3. RAE analysis of well-differentiated liposarcoma Model Chromosome Region start Region end Size q value freqX0* # genes Genes Amp 1 145009467 145122002 112536 0.097 21.8 2 PRKAB2,PDIA3P Amp 1 145224467 146188434 963968 0.029 23.6 10 CHD1L,BCL9,ACP6,GJA5,GJA8,GPR89B,GPR89C,PDZK1P1,RP11-94I2.2,NBPF11 Amp 1 147475854 148412469 936616 0.034 23.6 20 PPIAL4A,FCGR1A,HIST2H2BF,HIST2H3D,HIST2H2AA4,HIST2H2AA3,HIST2H3A,HIST2H3C,HIST2H4B,HIST2H4A,HIST2H2BE, HIST2H2AC,HIST2H2AB,BOLA1,SV2A,SF3B4,MTMR11,OTUD7B,VPS45,PLEKHO1 Amp 1 148582896 153398462 4815567 1.5E-05 49.1 152 PRPF3,RPRD2,TARS2,ECM1,ADAMTSL4,MCL1,ENSA,GOLPH3L,HORMAD1,CTSS,CTSK,ARNT,SETDB1,LASS2,ANXA9, FAM63A,PRUNE,BNIPL,C1orf56,CDC42SE1,MLLT11,GABPB2,SEMA6C,TNFAIP8L2,LYSMD1,SCNM1,TMOD4,VPS72, PIP5K1A,PSMD4,ZNF687,PI4KB,RFX5,SELENBP1,PSMB4,POGZ,CGN,TUFT1,SNX27,TNRC4,MRPL9,OAZ3,TDRKH,LINGO4, RORC,THEM5,THEM4,S100A10,S100A11,TCHHL1,TCHH,RPTN,HRNR,FLG,FLG2,CRNN,LCE5A,CRCT1,LCE3E,LCE3D,LCE3C,LCE3B, LCE3A,LCE2D,LCE2C,LCE2B,LCE2A,LCE4A,KPRP,LCE1F,LCE1E,LCE1D,LCE1C,LCE1B,LCE1A,SMCP,IVL,SPRR4,SPRR1A,SPRR3, SPRR1B,SPRR2D,SPRR2A,SPRR2B,SPRR2E,SPRR2F,SPRR2C,SPRR2G,LELP1,LOR,PGLYRP3,PGLYRP4,S100A9,S100A12,S100A8, S100A7A,S100A7L2,S100A7,S100A6,S100A5,S100A4,S100A3,S100A2,S100A16,S100A14,S100A13,S100A1,C1orf77,SNAPIN,ILF2, NPR1,INTS3,SLC27A3,GATAD2B,DENND4B,CRTC2,SLC39A1,CREB3L4,JTB,RAB13,RPS27,NUP210L,TPM3,C1orf189,C1orf43,UBAP2L,HAX1, AQP10,ATP8B2,IL6R,SHE,TDRD10,UBE2Q1,CHRNB2,ADAR,KCNN3,PMVK,PBXIP1,PYGO2,SHC1,CKS1B,FLAD1,LENEP,ZBTB7B,DCST2, DCST1,ADAM15,EFNA4,EFNA3,EFNA1,RAG1AP1,DPM3 Amp 1 -

Canonical WNT Signalling Determines Lineage Specificity in Wilms Tumour
Oncogene (2009) 28, 1063–1075 & 2009 Macmillan Publishers Limited All rights reserved 0950-9232/09 $32.00 www.nature.com/onc ORIGINAL ARTICLE Canonical WNT signalling determines lineage specificity in Wilms tumour R Fukuzawa, MR Anaka, RJ Weeks, IM Morison and AE Reeve Cancer Genetics Laboratory, Department of Biochemistry, University of Otago, Dunedin, New Zealand Wilms tumours (WTs) have two distinct types of histology to translocation of the protein to the nucleus (Clevers, with or without ectopic mesenchymal elements, suggesting 2006). Nuclear accumulation of b-catenin is therefore a that WTs arise from either the mesenchymal or epithelial surrogate marker for activation of the WNT-signalling nephrogenic lineages. Regardless of the presence or pathway in many types of cancers (Clevers, 2006). absence of CTNNB1 mutations, nuclear accumulation of Wilms tumour (WT) is an embryonal tumour that b-catenin is often observed in WTs with ectopic mesench- recapitulates kidney development. WT typically has a ymal elements. Here, we addressed the relationship triphasic histology, which consists of varying propor- between the WNT-signalling pathway and lineage in tions of blastemal, epithelial and stromal elements WTs by examining CTNNB1 and WT1 mutations, (Beckwith et al., 1990). The stromal tissues occasionally nuclear accumulation of b-catenin, tumour histology and contain ectopic mesenchymal elements such as rhabdo- gene expression profiles. In addition, we screened for myoblasts, chondrocytes and adipocytes. Remarkably, mutations in WTX, which has been proposed to be a the presence or absence of ectopic mesenchymal negative regulator of the canonical WNT-signalling elements in WT histology is associated with specific pathway. Unsupervised clustering analysis identified two precursor lesions, intralobar and perilobar nephrogenic classes of tumours: mesenchymal lineage WNT-dependent rests (ILNR and PLNR), respectively (Beckwith et al., tumours, and epithelial lineage WNT-independent tu- 1990). -

Developmental Neurobehavioral Toxicity of Bisphenol a in Zebrafish (Danio Rerio)
AN ABSTRACT OF THE DISSERTATION OF Katerine Schletz Saili for the degree of Doctor of Philosophy in Toxicology presented on November 13, 2012. Title: Developmental Neurobehavioral Toxicity of Bisphenol A in Zebrafish (Danio rerio) Abstract approved: _____________________________________________________ Robert L. Tanguay Billions of pounds of bisphenol A (BPA) are produced annually around the globe for the manufacture of numerous consumer products, including polycarbonate food and water containers, the protective resin linings of food cans, thermal printing paper, and dental fillings. BPA exposure during nervous system development has been associated with learning and behavioral impairments in animal models. The mode of action for these effects is not clearly defined. While BPA is a weak estrogen receptor (ER) agonist, it is also an estrogen-related receptor gamma (ERRγ) agonist. ERRγ binds BPA with 100 times greater affinity than ERs. This study was designed to test the hypothesis that exposure to human-relevant BPA concentrations impacts nervous system development and behavior through ERRγ activation. To examine whether BPA behaves more like an ER or ERRγ ligand, two positive control compounds were used throughout the study: 17β-estradiol (E2) and GSK4716, ER and ERRγ agonists, respectively. Initial behavior testing results included the observation that neurodevelopmental exposure to 0.01 or 0.1 μM BPA led to hyperactivity in larvae, while exposure to 0.1 or 1 μM BPA led to learning deficits in adult zebrafish. Exposure to 0.1 μM E2 or GSK4716 also led to larval hyperactivity. To identify early molecular signaling events that lead to the observed neurobehavioral phenotypes, a global gene expression analysis using a 135K zebrafish microarray was conducted.