Cholinesterase-Like Domains in Enzymes and Structural Proteins
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rnase 2 Sirna (H): Sc-92235
SANTA CRUZ BIOTECHNOLOGY, INC. RNase 2 siRNA (h): sc-92235 BACKGROUND PRODUCT RNase 2 [ribonuclease, RNase A family, 2 (liver, eosinophil-derived neuro- RNase 2 siRNA (h) is a pool of 2 target-specific 19-25 nt siRNAs designed toxin)], also known as non-secretory ribonuclease, EDN (eosinophil-derived to knock down gene expression. Each vial contains 3.3 nmol of lyophilized neurotoxin), RNase UpI-2 or RNS2, is a 161 amino acid protein that belongs siRNA, sufficient for a 10 µM solution once resuspended using protocol to the pancreatic ribonuclease family. Localizing to lysosome and cytoplasmic below. Suitable for 50-100 transfections. Also see RNase 2 shRNA granules, RNase 2 is expressed in leukocytes, liver, spleen, lung and body Plasmid (h): sc-92235-SH and RNase 2 shRNA (h) Lentiviral Particles: fluids. RNase 2 functions as a pyrimidine specific nuclease, and has a slight sc-92235-V as alternate gene silencing products. preference for uracil. RNase 2 is capable of various biological activities, For independent verification of RNase 2 (h) gene silencing results, we including mediation of chemotactic activity and endonucleolytic cleavage of also provide the individual siRNA duplex components. Each is available as nucleoside 3'-phosphates and 3'-phosphooligonucleotides. The gene encoding 3.3 nmol of lyophilized siRNA. These include: sc-92235A and sc-92235B. RNase 2 maps to human chromosome 14q11.2. STORAGE AND RESUSPENSION REFERENCES Store lyophilized siRNA duplex at -20° C with desiccant. Stable for at least 1. Yasuda, T., Sato, W., Mizuta, K. and Kishi, K. 1988. Genetic polymorphism one year from the date of shipment. -

GRAS Notice 653, Lysophospholipase from Aspergillus Nishimurae
GRAS Notice (GRN) No. 653 http://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/NoticeInventory/default.htm ORIGINAL SUBMISSION 000001 AB Enzymes GmbH – Feldbergstrasse 78 , D-64293 Darmstadt May 5, 2016 Office of Food Additive Safety (HFS-255), Center for Food Safety and Applied Nutrition, Food and Drug Administration, 5100 Paint Branch Parkway, College Park, MD 20740. RE: GRAS NOTICE FOR lysophospholipase Enzyme Preparation From Trichoderma reesei Pursuant to proposed 21 C.F.R § 170.36, AB Enzymes GmbH is providing in electronic media format (determined to be free of computer viruses), based on scientific procedures – a generally recognized as safe (GRAS) notification for lysophospholipase enzyme preparation from Trichoderma reesei for use as a processing aid used in starch processing. The lysophospholipase enzyme preparation described herein when used as described above and in the attached GRAS notice is exempt from the premarket approval requirements applicable to food additives set forth in Section 409 of the Food, Drug, and Cosmetic Act and corresponding regulations. Please contact the undersigned by telephone or email if you have any questions or additional information is required. Candice Cryne Regulatory Affairs Manager 1 647-919-3964 [email protected] 000002 ~ AB Enzymes AB Enzymes GmbH - Feldbergstrasse 78, 0 -64293 Darmstadt ... AUf'~- 3 GR ·N000(,5 {~g~~~\§\Eli] May 5, 2016 Mfl.'< 21 201S Office of Food Additive Safety (HFS-255), OFFICE OF Center for Food Safety and Applied Nutrition, FOOD ADDITIVE SAFEi'f l Food and Drug Administration, L 5100 Paint Branch Parkway, College Park, MD 20740. RE: GRAS NOTICE FOR lysophospholipase Enzyme Preparation From Trichoderma reesei Pursuant to proposed 21 C.F.R § 170.36, AB Enzymes GmbH is providing in electronic media format (determined to be free of computer viruses), based on scientific procedures- a generally recognized as safe (GRAS) notification for lysophospholipase enzyme preparation from Trichoderma reesei for use as a processing aid used in starch processing . -

Cutinases from Mycobacterium Tuberculosis
Identification of Residues Involved in Substrate Specificity and Cytotoxicity of Two Closely Related Cutinases from Mycobacterium tuberculosis Luc Dedieu, Carole Serveau-Avesque, Ste´phane Canaan* CNRS - Aix-Marseille Universite´ - Enzymologie Interfaciale et Physiologie de la Lipolyse - UMR 7282, Marseille, France Abstract The enzymes belonging to the cutinase family are serine enzymes active on a large panel of substrates such as cutin, triacylglycerols, and phospholipids. In the M. tuberculosis H37Rv genome, seven genes coding for cutinase-like proteins have been identified with strong immunogenic properties suggesting a potential role as vaccine candidates. Two of these enzymes which are secreted and highly homologous, possess distinct substrates specificities. Cfp21 is a lipase and Cut4 is a phospholipase A2, which has cytotoxic effects on macrophages. Structural overlay of their three-dimensional models allowed us to identify three areas involved in the substrate binding process and to shed light on this substrate specificity. By site-directed mutagenesis, residues present in these Cfp21 areas were replaced by residues occurring in Cut4 at the same location. Three mutants acquired phospholipase A1 and A2 activities and the lipase activities of two mutants were 3 and 15 fold greater than the Cfp21 wild type enzyme. In addition, contrary to mutants with enhanced lipase activity, mutants that acquired phospholipase B activities induced macrophage lysis as efficiently as Cut4 which emphasizes the relationship between apparent phospholipase A2 activity and cytotoxicity. Modification of areas involved in substrate specificity, generate recombinant enzymes with higher activity, which may be more immunogenic than the wild type enzymes and could therefore constitute promising candidates for antituberculous vaccine production. -
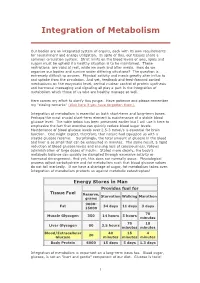
Integration of Metabolism
Integration of Metabolism Our bodies are an integrated system of organs, each with its own requirements for nourishment and energy utilization. In spite of this, our tissues share a common circulation system. Strict limits on the blood levels of ions, lipids and sugars must be upheld if a healthy situation is to be maintained. These restrictions are valid at rest, while we work and after meals. How do we organize our bodies and survive under differing situations? The question is extremely difficult to answer. Physical activity and meals greatly alter influx to and uptake from the circulation. And yet, feedback and feed-forward control mechanisms on the enzymatic level, central nuclear control of protein synthesis and hormonal messaging and signaling all play a part in the integration of metabolism which those of us who are healthy manage so well. Here comes my effort to clarify this jungle. Have patience and please remember my "closing remarks" (click here if you have forgotten them). Integration of metabolism is essential on both short-term and long-term bases. Perhaps the most crucial short-term element is maintenance of a stable blood glucose level. The table below has been presented earlier but I will use it here to emphasize the fact that exercise can quickly reduce blood sugar levels. Maintenance of blood glucose levels over 2.5-3 mmol/s is essential for brain function. One might expect, therefore, that nature had equipped us with a sizable glucose reserve. Surprisingly, the total amount of glucose in the blood and liver is so small that can be exhausted in minutes. -

Universal Riboclone™ Cdna Synthesis System
TECHNICAL MANUAL Universal RiboClone™ cDNA Synthesis System Instructions for Use of Product C4360 Revised 4/21 TM038 Universal RiboClone™ cDNA Synthesis System All technical literature is available at: www.promega.com/protocols/ Visit the web site to verify that you are using the most current version of this Technical Manual. E-mail Promega Technical Services if you have questions on use of this system: [email protected] 1. Description .........................................................................................................................................2 2. Product Components and Storage Conditions ........................................................................................3 3. General Considerations .......................................................................................................................4 3.A. Methods of cDNA Synthesis .........................................................................................................4 3.B. Choice of Primers .......................................................................................................................4 3.C. cDNA Cloning ............................................................................................................................4 3.D. Choice of Vector .........................................................................................................................8 3.E. RNA Preparation and Handling ...................................................................................................8 -

The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z
REVIEW pubs.acs.org/CR The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease Jonathan Z. Long* and Benjamin F. Cravatt* The Skaggs Institute for Chemical Biology and Department of Chemical Physiology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States CONTENTS 2.4. Other Phospholipases 6034 1. Introduction 6023 2.4.1. LIPG (Endothelial Lipase) 6034 2. Small-Molecule Hydrolases 6023 2.4.2. PLA1A (Phosphatidylserine-Specific 2.1. Intracellular Neutral Lipases 6023 PLA1) 6035 2.1.1. LIPE (Hormone-Sensitive Lipase) 6024 2.4.3. LIPH and LIPI (Phosphatidic Acid-Specific 2.1.2. PNPLA2 (Adipose Triglyceride Lipase) 6024 PLA1R and β) 6035 2.1.3. MGLL (Monoacylglycerol Lipase) 6025 2.4.4. PLB1 (Phospholipase B) 6035 2.1.4. DAGLA and DAGLB (Diacylglycerol Lipase 2.4.5. DDHD1 and DDHD2 (DDHD Domain R and β) 6026 Containing 1 and 2) 6035 2.1.5. CES3 (Carboxylesterase 3) 6026 2.4.6. ABHD4 (Alpha/Beta Hydrolase Domain 2.1.6. AADACL1 (Arylacetamide Deacetylase-like 1) 6026 Containing 4) 6036 2.1.7. ABHD6 (Alpha/Beta Hydrolase Domain 2.5. Small-Molecule Amidases 6036 Containing 6) 6027 2.5.1. FAAH and FAAH2 (Fatty Acid Amide 2.1.8. ABHD12 (Alpha/Beta Hydrolase Domain Hydrolase and FAAH2) 6036 Containing 12) 6027 2.5.2. AFMID (Arylformamidase) 6037 2.2. Extracellular Neutral Lipases 6027 2.6. Acyl-CoA Hydrolases 6037 2.2.1. PNLIP (Pancreatic Lipase) 6028 2.6.1. FASN (Fatty Acid Synthase) 6037 2.2.2. PNLIPRP1 and PNLIPR2 (Pancreatic 2.6.2. -

Diagnostic Value of Serum Enzymes-A Review on Laboratory Investigations
Review Article ISSN 2250-0480 VOL 5/ ISSUE 4/OCT 2015 DIAGNOSTIC VALUE OF SERUM ENZYMES-A REVIEW ON LABORATORY INVESTIGATIONS. 1VIDYA SAGAR, M.SC., 2DR. VANDANA BERRY, MD AND DR.ROHIT J. CHAUDHARY, MD 1Vice Principal, Institute of Allied Health Sciences, Christian Medical College, Ludhiana 2Professor & Ex-Head of Microbiology Christian Medical College, Ludhiana 3Assistant Professor Department of Biochemistry Christian Medical College, Ludhiana ABSTRACT Enzymes are produced intracellularly, and released into the plasma and body fluids, where their activities can be measured by their abilities to accelerate the particular chemical reactions they catalyze. But different serum enzymes are raised when different tissues are damaged. So serum enzyme determination can be used both to detect cellular damage and to suggest its location in situ. Some of the biochemical markers such as alanine aminotransferase, aspartate aminotransferase, alkaline phasphatase, gamma glutamyl transferase, nucleotidase, ceruloplasmin, alpha fetoprotein, amylase, lipase, creatine phosphokinase and lactate dehydrogenase are mentioned to evaluate diseases of liver, pancreas, skeletal muscle, bone, etc. Such enzyme test may assist the physician in diagnosis and treatment. KEYWORDS: Liver Function tests, Serum Amylase, Lipase, CPK and LDH. INTRODUCTION mitochondrial AST is seen in extensive tissue necrosis during myocardial infarction and also in chronic Liver diseases like liver tissue degeneration DIAGNOSTIC SERUM ENZYME and necrosis². But lesser amounts are found in Enzymes are very helpful in the diagnosis of brain, pancreas and lung. Although GPT is plentiful cardiac, hepatic, pancreatic, muscular, skeltal and in the liver and occurs only in the small amount in malignant disorders. Serum for all enzyme tests the other tissues. -

GC-MS Metabolic Profile and -Glucosidase-, -Amylase-, Lipase-, and Acetylcholinesterase-Inhibitory Activities of Eight Peach
molecules Article GC-MS Metabolic Profile and α-Glucosidase-, α-Amylase-, Lipase-, and Acetylcholinesterase-Inhibitory Activities of Eight Peach Varieties Dasha Mihaylova 1,* , Ivelina Desseva 2,* , Aneta Popova 3 , Ivayla Dincheva 4 , Radka Vrancheva 2 , Anna Lante 5 and Albert Krastanov 1 1 Department of Biotechnology, Technological Faculty, University of Food Technologies, 4002 Plovdiv, Bulgaria; [email protected] 2 Department of Analytical Chemistry and Physical Chemistry, Technological Faculty, University of Food Technologies, 4002 Plovdiv, Bulgaria; [email protected] 3 Department of Catering and Tourism, Economics Faculty, University of Food Technologies, 4002 Plovdiv, Bulgaria; [email protected] 4 AgroBioInstitute, Agricultural Academy, 8 Dr. Tsankov Blvd., 1164 Sofia, Bulgaria; [email protected] 5 Department of Agronomy, Food, Natural Resources, Animals, and Environment—DAFNAE, Agripolis, University of Padova, 35020 Legnaro, Italy; [email protected] * Correspondence: [email protected] (D.M.); [email protected] (I.D.) Abstract: The inhibition of certain digestive enzymes by target food matrices represents a new approach in the treatment of socially significant diseases. Proving the ability of fruits to inhibit such enzymes can support the inclusion of specific varieties in the daily diets of patients with diabetes, obesity, Alzheimer’s disease, etc., providing them with much more than just valuable Citation: Mihaylova, D.; Desseva, I.; micro- and macromolecules. The current study aimed atidentifying and comparing the GC-MS Popova, A.; Dincheva, I.; Vrancheva, metabolic profiles of eight peach varieties (“Filina”, “Ufo 4, “Gergana”, “Laskava”, “July Lady”, R.; Lante, A.; Krastanov, A. GC-MS “Flat Queen”, “Evmolpiya”, and “Morsiani 90”) grown in Bulgaria (local and introduced) and to Metabolic Profile and α-Glucosidase-, evaluate the inhibitory potential of their extracts towards α-glucosidase, α-amylase, lipase, and α-Amylase-, Lipase-, and acetylcholinesterase. -

(10) Patent No.: US 8119385 B2
US008119385B2 (12) United States Patent (10) Patent No.: US 8,119,385 B2 Mathur et al. (45) Date of Patent: Feb. 21, 2012 (54) NUCLEICACIDS AND PROTEINS AND (52) U.S. Cl. ........................................ 435/212:530/350 METHODS FOR MAKING AND USING THEMI (58) Field of Classification Search ........................ None (75) Inventors: Eric J. Mathur, San Diego, CA (US); See application file for complete search history. Cathy Chang, San Diego, CA (US) (56) References Cited (73) Assignee: BP Corporation North America Inc., Houston, TX (US) OTHER PUBLICATIONS c Mount, Bioinformatics, Cold Spring Harbor Press, Cold Spring Har (*) Notice: Subject to any disclaimer, the term of this bor New York, 2001, pp. 382-393.* patent is extended or adjusted under 35 Spencer et al., “Whole-Genome Sequence Variation among Multiple U.S.C. 154(b) by 689 days. Isolates of Pseudomonas aeruginosa” J. Bacteriol. (2003) 185: 1316 1325. (21) Appl. No.: 11/817,403 Database Sequence GenBank Accession No. BZ569932 Dec. 17. 1-1. 2002. (22) PCT Fled: Mar. 3, 2006 Omiecinski et al., “Epoxide Hydrolase-Polymorphism and role in (86). PCT No.: PCT/US2OO6/OOT642 toxicology” Toxicol. Lett. (2000) 1.12: 365-370. S371 (c)(1), * cited by examiner (2), (4) Date: May 7, 2008 Primary Examiner — James Martinell (87) PCT Pub. No.: WO2006/096527 (74) Attorney, Agent, or Firm — Kalim S. Fuzail PCT Pub. Date: Sep. 14, 2006 (57) ABSTRACT (65) Prior Publication Data The invention provides polypeptides, including enzymes, structural proteins and binding proteins, polynucleotides US 201O/OO11456A1 Jan. 14, 2010 encoding these polypeptides, and methods of making and using these polynucleotides and polypeptides. -

The Role of Phospholipase D (Pld) and Grb2 in Chemotaxis
THE ROLE OF PHOSPHOLIPASE D (PLD) AND GRB2 IN CHEMOTAXIS A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By KATIE J. KNAPEK B.S., Indiana University of Pennyslvania, 2006 2008 Wright State University WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES December 19, 2008 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISON BY Katie J. Knapek ENTITLED The Role of Phospholipase D (PLD) and Grb2 in Chemotaxis BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science. _____________________________ Julian Gomez-Cambronero, Ph. D. Thesis Director _____________________________ Barbara Hull, Ph. D. Program Director Committee on Final Examination _______________________ Julian Gomez-Cambronero, Ph. D. _______________________ Nancy Bigley, Ph. D. _______________________ Mill Miller, Ph. D. ________________________ Joseph F. Thomas Jr., Ph. D. Dean, School of Graduate Studies ABSTRACT Knapek, Katie J. M.S., Department of Biological Sciences, Microbiology and Immunology Program, Wright State University, 2008. The Role of Phospholipase D (PLD) and Grb2 in Chemotaxis. Phospholipase D (PLD) is an enzyme that hydrolyzes phosphatidylcholine yielding choline and phosphatidic acid. PLD is activated by mitogens (lead to cell division) and motogens (leading to cell migration). PLD is known to contribute to cellular proliferation and deregulated expression of PLD has been implicated in several human cancers. PLD has been found to play a role in leukocyte chemotaxis and adhesion as studied through the formation of chemokine gradients. We have established a model of cell migration comprising three cell lines: macrophages RAW 264.7 and LR-5 (for innate defense), and fibroblast COS-7 cells (for wound healing). -

Multiple Roles of Phosphoinositide-Specific Phospholipase C Isozymes
BMB reports Mini Review Multiple roles of phosphoinositide-specific phospholipase C isozymes Pann-Ghill Suh1,*, Jae-Il Park1, Lucia Manzoli2, Lucio Cocco2, Joanna C. Peak3, Matilda Katan3, Kiyoko Fukami4, Tohru Kataoka5, Sanguk Yun1 & Sung Ho Ryu1 1Division of Molecular and Life Sciences, Pohang University of Science and Technology, Pohang, Korea, 2Cellular Signaling Laboratory, Department of Anatomical Sciences, University of Bologna, Via Irnerio, 48 I-40126, Bologna, Italy, 3Cancer Research UK Centre for Cell and Molecular Biology, Chester Beatty Laboratories, The Institute of Cancer Research, Fulham Road, London SW3 6JB, UK, 4Laboratory of Genome and Biosignal, Tokyo University of Pharmacy and Life Science, 1432-1 Horinouchi, Hachioji, 192-0392 Tokyo, Japan, 5Division of Molecular Biology, Department of Biochemistry and Molecular Biology, Kobe University Graduate School of Medicine, Chuo-ku, Kobe, Japan Phosphoinositide-specific phospholipase C is an effector mole- cellular calcium release (1-3; Fig. 1a). cule in the signal transduction process. It generates two sec- The first evidence of PLC activity was suggested by Hokin et ond messengers, inositol-1,4,5-trisphosphate and diacylglycer- al. in 1953 who reported specific hydrolysis of phospholipids ol from phosphatidylinositol 4,5-bisphosphate. Currently, thir- in pigeon’s pancreas slices after cholinergic stimulation (4). teen mammal PLC isozymes have been identified, and they are The authors showed that the enhanced turnover of phosphor- divided into six groups: PLC-β, -γ, -δ, -ε, -ζ and -η. Sequence ylinositol groups of phosphatidylinositol occurred in cells as a analysis studies demonstrated that each isozyme has more response to a variety of stimuli. In 1983, Streb et al. -

Pancreatic Amylase and Lipase Plasma Concentrations Are
Diabetes Care Volume 38, May 2015 e71 Pancreatic Amylase and Lipase Plasma David P. Sonne,1 Tina Vilsbøll,1 and Filip K. Knop1,2 Concentrations Are Unaffected by Increments in Endogenous GLP-1 Levels Following Liquid Meal Tests Diabetes Care 2015;38:e71–e72 | DOI: 10.2337/dc14-2751 Recent findings have suggested that amylase and lipase in plasma following patients with type 2 diabetes (P , incretin-based therapies promote pan- the ingestion of oral glucose and three 0.05), whereas lipase concentrations creatic inflammation and possibly cell isocaloric and isovolemic liquid mealsd were similar. Neither of the enzymes proliferation within the endocrine and all of which exerted normal endogenous increased following nutrient ingestion, exocrine pancreas (1). However, these GLP-1 secretion (4)din patients with suggesting that postprandial elevations studies have been met with substan- type 2 diabetes and matched control of endogenous GLP-1 (two- to three- tive criticism based on technical and subjects. fold) cannot trigger enzyme release methodical issues (2). Nevertheless, Detailed description of the experimen- from the human pancreas, at least not incretin-based therapies seem to result tal procedures and subjects was provided acutely. in small increases (within normal range) previously (4). In short, pancreas-specific These results suggest that the obser- in plasma concentrations of amylase amylase and lipase concentrations were vation of elevated plasma amylase and andlipaseinpatientsreceivingthese measured in plasma from 15 patients lipase