Tet Proteins Influence the Balance Between PNAS PLUS Neuroectodermal and Mesodermal Fate Choice by Inhibiting Wnt Signaling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

3 Embryology and Development
BIOL 6505 − INTRODUCTION TO FETAL MEDICINE 3. EMBRYOLOGY AND DEVELOPMENT Arlet G. Kurkchubasche, M.D. INTRODUCTION Embryology – the field of study that pertains to the developing organism/human Basic embryology –usually taught in the chronologic sequence of events. These events are the basis for understanding the congenital anomalies that we encounter in the fetus, and help explain the relationships to other organ system concerns. Below is a synopsis of some of the critical steps in embryogenesis from the anatomic rather than molecular basis. These concepts will be more intuitive and evident in conjunction with diagrams and animated sequences. This text is a synopsis of material provided in Langman’s Medical Embryology, 9th ed. First week – ovulation to fertilization to implantation Fertilization restores 1) the diploid number of chromosomes, 2) determines the chromosomal sex and 3) initiates cleavage. Cleavage of the fertilized ovum results in mitotic divisions generating blastomeres that form a 16-cell morula. The dense morula develops a central cavity and now forms the blastocyst, which restructures into 2 components. The inner cell mass forms the embryoblast and outer cell mass the trophoblast. Consequences for fetal management: Variances in cleavage, i.e. splitting of the zygote at various stages/locations - leads to monozygotic twinning with various relationships of the fetal membranes. Cleavage at later weeks will lead to conjoined twinning. Second week: the week of twos – marked by bilaminar germ disc formation. Commences with blastocyst partially embedded in endometrial stroma Trophoblast forms – 1) cytotrophoblast – mitotic cells that coalesce to form 2) syncytiotrophoblast – erodes into maternal tissues, forms lacunae which are critical to development of the uteroplacental circulation. -

The Zebrafish Presomitic Mesoderm Elongates Through Compression-Extension
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.11.434927; this version posted March 11, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. The zebrafish presomitic mesoderm elongates through compression-extension Lewis Thomson1, Leila Muresan2 and Benjamin Steventon1* 1) Department of Genetics, University of Cambridge, Cambridge, UK, CB2 3EH 2) Cambridge Advanced Imaging Centre, University of Cambridge, Cambridge, UK *[email protected] Abstract In vertebrate embryos the presomitic mesoderm become progressively segmented into somites at the anterior end while extending along the anterior- posterior axis. A commonly adopted model to explain how this tissue elongates is that of posterior growth, driven in part by the addition of new cells from uncommitted progenitor populations in the tailbud. However, in zebrafish, much of somitogenesis is associated with an absence of overall volume increase and posterior progenitors do not contribute new cells until the final stages of somitogenesis. Here, we perform a comprehensive 3D morphometric analysis of the paraxial mesoderm and reveal that extension is linked to a volumetric decrease, compression in both dorsal-ventral and medio-lateral axes, and an increase in cell density. We also find that individual cells decrease in their cell volume over successive somite stages. Live cell tracking confirms that much of this tissue deformation occurs within the presomitic mesoderm progenitor zone and is associated with non-directional rearrangement. Furthermore, unlike the trunk somites that are laid down during gastrulation, tail somites develop from a tissue that can continue to elongate in the absence of functional PCP signalling. -

Theory and Practice of Lineage Tracing
REGENERATIVE MEDICINE Theory and Practice of Lineage Tracing a,b YA-CHIEH HSU Key Words. Cell surface markers • Stem cell-microenvironment interactions • Cell cycle • Cell biology • Adult stem cells aDepartment of Stem Cell ABSTRACT and Regenerative Biology, Lineage tracing is a method that delineates all progeny produced by a single cell or a group of Harvard University, cells. The possibility of performing lineage tracing initiated the field of Developmental Biology Cambridge, Massachusetts, and continues to revolutionize Stem Cell Biology. Here, I introduce the principles behind a suc- USA; bHarvard Stem Cell cessful lineage-tracing experiment. In addition, I summarize and compare different methods for Institute, Cambridge, conducting lineage tracing and provide examples of how these strategies can be implemented Massachusetts, USA to answer fundamental questions in development and regeneration. The advantages and limita- Correspondence: Ya-Chieh Hsu, tions of each method are also discussed. STEM CELLS 2015;33:3197–3204 Department of Stem Cell and Regenerative Biology, Harvard University, 7 Divinity Ave., SIGNIFICANCE STATEMENT Cambridge, Massachusetts 02138, USA. Telephone: Lineage relationships allow one to understand the cellular hierarchy of a tissue or organ, which (617) 496-3757; Fax: (617) 496- is central to both Developmental Biology and Stem Cell Biology. Different approaches have 3763; e-mail: yachiehhsu@fas. been developed to conduct lineage tracing. Understanding the underlying basis of these harvard.edu approaches, as well as their advantages and limitations, will allow one to choose the most Received March 26, 2015; appropriate method for a given question. accepted for publication June 23, 2015; first published online STEM CELLS in EXPRESS August 3, cells that are marked at the initial time point, 2015. -

From Bipotent Neuromesodermal Progenitors to Neural-Mesodermal Interactions During Embryonic Development
International Journal of Molecular Sciences Review From Bipotent Neuromesodermal Progenitors to Neural-Mesodermal Interactions during Embryonic Development Nitza Kahane and Chaya Kalcheim * Department of Medical Neurobiology, Institute of Medical Research Israel-Canada (IMRIC) and the Edmond and Lily Safra Center for Brain Sciences (ELSC), Hebrew University of Jerusalem-Hadassah Medical School, P.O. Box 12272, Jerusalem 9112102, Israel; [email protected] * Correspondence: [email protected] Abstract: To ensure the formation of a properly patterned embryo, multiple processes must operate harmoniously at sequential phases of development. This is implemented by mutual interactions between cells and tissues that together regulate the segregation and specification of cells, their growth and morphogenesis. The formation of the spinal cord and paraxial mesoderm derivatives exquisitely illustrate these processes. Following early gastrulation, while the vertebrate body elongates, a pop- ulation of bipotent neuromesodermal progenitors resident in the posterior region of the embryo generate both neural and mesodermal lineages. At later stages, the somitic mesoderm regulates aspects of neural patterning and differentiation of both central and peripheral neural progenitors. Reciprocally, neural precursors influence the paraxial mesoderm to regulate somite-derived myogen- esis and additional processes by distinct mechanisms. Central to this crosstalk is the activity of the axial notochord, which, via sonic hedgehog signaling, plays pivotal roles in neural, skeletal muscle and cartilage ontogeny. Here, we discuss the cellular and molecular basis underlying this complex Citation: Kahane, N.; Kalcheim, C. developmental plan, with a focus on the logic of sonic hedgehog activities in the coordination of the From Bipotent Neuromesodermal Progenitors to Neural-Mesodermal neural-mesodermal axis. -

The Genetic Basis of Mammalian Neurulation
REVIEWS THE GENETIC BASIS OF MAMMALIAN NEURULATION Andrew J. Copp*, Nicholas D. E. Greene* and Jennifer N. Murdoch‡ More than 80 mutant mouse genes disrupt neurulation and allow an in-depth analysis of the underlying developmental mechanisms. Although many of the genetic mutants have been studied in only rudimentary detail, several molecular pathways can already be identified as crucial for normal neurulation. These include the planar cell-polarity pathway, which is required for the initiation of neural tube closure, and the sonic hedgehog signalling pathway that regulates neural plate bending. Mutant mice also offer an opportunity to unravel the mechanisms by which folic acid prevents neural tube defects, and to develop new therapies for folate-resistant defects. 6 ECTODERM Neurulation is a fundamental event of embryogenesis distinct locations in the brain and spinal cord .By The outer of the three that culminates in the formation of the neural tube, contrast, the mechanisms that underlie the forma- embryonic (germ) layers that which is the precursor of the brain and spinal cord. A tion, elevation and fusion of the neural folds have gives rise to the entire central region of specialized dorsal ECTODERM, the neural plate, remained elusive. nervous system, plus other organs and embryonic develops bilateral neural folds at its junction with sur- An opportunity has now arisen for an incisive analy- structures. face (non-neural) ectoderm. These folds elevate, come sis of neurulation mechanisms using the growing battery into contact (appose) in the midline and fuse to create of genetically targeted and other mutant mouse strains NEURAL CREST the neural tube, which, thereafter, becomes covered by in which NTDs form part of the mutant phenotype7.At A migratory cell population that future epidermal ectoderm. -

Delta-Notch Signaling: the Long and the Short of a Neuron’S Influence on Progenitor Fates
Journal of Developmental Biology Review Delta-Notch Signaling: The Long and the Short of a Neuron’s Influence on Progenitor Fates Rachel Moore 1,* and Paula Alexandre 2,* 1 Centre for Developmental Neurobiology, King’s College London, London SE1 1UL, UK 2 Developmental Biology and Cancer, University College London Great Ormond Street Institute of Child Health, London WC1N 1EH, UK * Correspondence: [email protected] (R.M.); [email protected] (P.A.) Received: 18 February 2020; Accepted: 24 March 2020; Published: 26 March 2020 Abstract: Maintenance of the neural progenitor pool during embryonic development is essential to promote growth of the central nervous system (CNS). The CNS is initially formed by tightly compacted proliferative neuroepithelial cells that later acquire radial glial characteristics and continue to divide at the ventricular (apical) and pial (basal) surface of the neuroepithelium to generate neurons. While neural progenitors such as neuroepithelial cells and apical radial glia form strong connections with their neighbours at the apical and basal surfaces of the neuroepithelium, neurons usually form the mantle layer at the basal surface. This review will discuss the existing evidence that supports a role for neurons, from early stages of differentiation, in promoting progenitor cell fates in the vertebrates CNS, maintaining tissue homeostasis and regulating spatiotemporal patterning of neuronal differentiation through Delta-Notch signalling. Keywords: neuron; neurogenesis; neuronal apical detachment; asymmetric division; notch; delta; long and short range lateral inhibition 1. Introduction During the development of the central nervous system (CNS), neurons derive from neural progenitors and the Delta-Notch signaling pathway plays a major role in these cell fate decisions [1–4]. -

Molecular Cell and Developmental Biology Graduate Program 2018-2019
Molecular Cell and Developmental Biology Graduate Program 2018-2019 The Ph.D. track in Molecular Cell and Developmental Biology (MCD) is designed to prepare students for productive careers in biological research and teaching. This training program emphasizes applying diverse approaches, including biochemistry, genetics, genomics, and imaging, to addressing critical questions in molecular, cellular, and developmental biology. Interdisciplinary research is encouraged and supported by a diverse group of faculty from the Departments of Molecular Cell & Developmental Biology (MCD), Chemistry & Biochemistry (Chem), Microbiology & Environmental Toxicology (METX), Biomolecular Engineering (BME), the Santa Cruz Institute for Particle Physics (Physics), and Ecology & Evolutionary Biology (EEB). MCD Faculty James Ackman Visualizing structured activity throughout the developing vertebrate brain Manny Ares Mechanisms and regulation of splicing machinery; structure and function of small RNAs Joshua Arribere Analyzing RNA quality control in C. elegans Needhi Bhalla Meiotic chromosome dynamics Hinrich Boeger Chromatin structure and gene regulation Barry Bowman Membrane biochemistry, genetics, and molecular biology Susan Carpenter Long non-coding RNAs in innate immunity Bin Chen Molecular control of neuronal identity and connectivity in mammalian brains David Feldheim Topographic mapping in vertebrates/CNS Grant Hartzog Chromatin and transcription Lindsay Hinck Molecular basis of neuronal chemotropism Melissa Jurica Structural approaches to large macromolecular -

Understanding Paraxial Mesoderm Development and Sclerotome Specification for Skeletal Repair Shoichiro Tani 1,2, Ung-Il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3
Tani et al. Experimental & Molecular Medicine (2020) 52:1166–1177 https://doi.org/10.1038/s12276-020-0482-1 Experimental & Molecular Medicine REVIEW ARTICLE Open Access Understanding paraxial mesoderm development and sclerotome specification for skeletal repair Shoichiro Tani 1,2, Ung-il Chung2,3, Shinsuke Ohba4 and Hironori Hojo2,3 Abstract Pluripotent stem cells (PSCs) are attractive regenerative therapy tools for skeletal tissues. However, a deep understanding of skeletal development is required in order to model this development with PSCs, and for the application of PSCs in clinical settings. Skeletal tissues originate from three types of cell populations: the paraxial mesoderm, lateral plate mesoderm, and neural crest. The paraxial mesoderm gives rise to the sclerotome mainly through somitogenesis. In this process, key developmental processes, including initiation of the segmentation clock, formation of the determination front, and the mesenchymal–epithelial transition, are sequentially coordinated. The sclerotome further forms vertebral columns and contributes to various other tissues, such as tendons, vessels (including the dorsal aorta), and even meninges. To understand the molecular mechanisms underlying these developmental processes, extensive studies have been conducted. These studies have demonstrated that a gradient of activities involving multiple signaling pathways specify the embryonic axis and induce cell-type-specific master transcription factors in a spatiotemporal manner. Moreover, applying the knowledge of mesoderm development, researchers have attempted to recapitulate the in vivo development processes in in vitro settings, using mouse and human PSCs. In this review, we summarize the state-of-the-art understanding of mesoderm development and in vitro modeling of mesoderm development using PSCs. We also discuss future perspectives on the use of PSCs to generate skeletal tissues for basic research and clinical applications. -

Cell Fate and Cell Morphogenesis in Higher Plants
Cell fate and cell morphogenesis in higher plants John W Schiefelbein University of Michigan, Ann Arbor, USA The differentiation of plant cells depends on the regulation of cell fate and cell morphogenesis. Recent studies have led to the identification of mutants and the cloning of genes that influence these processes. In several instances, the genes encode products with homeodomains or Myb or Myc DNA-binding domains. Current Opinion in Genetics and Development 1994, 4:647451 Introduction mutants led eo the early notion that iUVl influences the Gte of leaf cells. Recent studies have shed light on the In higher plants, the formation and differentiation of normaI role of KNY. In localization experiments, KNl cells occurs in an orderly manner, ofien near meristems, is undetectable in leaves, but it is abundant in the api- which are groups of dividing cells chat act like permanent cal meristem and the immature axes of vegetative and stem cell populations. Cell differentiation in plants dif- floral shoots [7]. Also, the dominant Knl-N2 mutation fers horn that in animals because, for the most part, of a has been found to cause ectopic expression, such that lack of the relative cell movements that are characteristic KZVZ transcripts and KNl protein are detected in lateral of animal development. Also, the rigid plant cell wall veins of developing leaves (in addition to the normal makes the control of cell shape a critical aspect of cell accumulation in the meristem), which correlates well differentiation in plants. with the alterations in leaf cell fite and knot forma- The fate of undifferentiated plant cells is influenced pri- tion associated with Krzl mutants [7]. -

Genes for Extracellular Matrix-Degrading Metalloproteinases and Their Inhibitor, TIMP, Are Expressed During Early Mammalian Development
Downloaded from genesdev.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Genes for extracellular matrix-degrading metalloproteinases and their inhibitor, TIMP, are expressed during early mammalian development Carol A. Brenner, 1 Richard R. Adler, 2 Daniel A. Rappolee, Roger A. Pedersen, and Zena Werb 3 Laboratory of Radiobiology and Environmental Health, and Department of Anatomy, University of California, San Francisco, California 94143-0750 USA Extracellular matrix (ECM) remodeling accompanies cell migration, cell--cell interactions, embryo expansion, uterine implantation, and tissue invasion during mammalian embryogenesis. We have found that mouse embryos secrete functional ECM-degrading metalloproteinases, including collagenase and stromelysin, that are inhibitable by the tissue inhibitor of metalloproteinases (TIMP) and that are regulated during peri-implantation development and endoderm differentiation, mRNA transcripts for collagenase, stromelysin, and TIMP were detected as maternal transcripts in the unfertilized egg, were present at the zygote and cleavage stages, and increased at the blastocyst stage and with endoderm differentiation. These data suggest that metalloproteinases function in cell-ECM interactions during growth, development, and implantation of mammalian embryos. [Key Words: Collagenase; stromelysin; polymerase chain reaction; tissue inhibitor of metalloproteinases; maternal mRNA; endoderm differentiation] January 31, 1989; revised version accepted March 15, 1989. The extracellular matrix (ECM) forms a substrate for cell 1988}. The activity of these proteinases is regulated by attachment and migration and directs cell form and the tissue inhibitor of metalloproteinases {TIMPI function through cell-ECM interactions. ECM macro- (Murphy et al. 1985; Herron et al. 1986a, b; Gavrilovic et molecules first appear in the mammalian embryo in the al. -
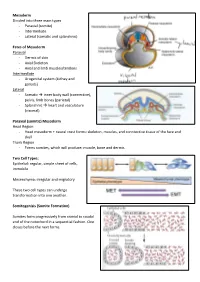
Mesoderm Divided Into Three Main Types - Paraxial (Somite) - Intermediate - Lateral (Somatic and Splanchnic)
Mesoderm Divided into three main types - Paraxial (somite) - Intermediate - Lateral (somatic and splanchnic) Fates of Mesoderm Paraxial - Dermis of skin - Axial Skeleton - Axial and limb muscles/tendons Intermediate - Urogenital system (kidney and gonads) Lateral - Somatic inner body wall (connective), pelvis, limb bones (parietal) - Splanchnic heart and vasculature (visceral) Paraxial (somitic) Mesoderm Head Region - Head mesoderm + neural crest forms: skeleton, muscles, and conntective tissue of the face and skull Trunk Region - Forms somites, which will produce: muscle, bone and dermis Two Cell Types: Epithelial: regular, simple sheet of cells, immobile Mesenchyma: irregular and migratory These two cell types can undergo transformation into one another. Somitogenisis (Somite Formation) Somites form progressively from cranial to caudal end of the notochord in a sequential fashion. One closes before the next forms. Somite Differentiation The somite splits into the epithelial dermamyotome (dermis/muscle) and the messenchymal sclerotome (skeletal). The somite is all paraxial mesoderm. Somite location determines the fate of its associates derma/myo/sclerotomes. Intermediate Mesoderm Urogenital system: - Kidneys - Gonads - Reproductive Duct Systems Runs alongside the paraxial mesoderm. Urogenital System Along mesonephric duct: - Pronephros, mesonephros, and metanephros - Pronephros fall away as gonad develops on ventral-medial side of mesonephros. - Metanephrogenic mesenchyme gives rise to kidney. The mesonephric duct will become the Wolffian duct forming at the nephric bud. The Mullerian duct forms via an invagination on the dorsal side of the nephric duct. The gonad will degenerate one of the two ducts depending on the hormones it produces. XX degenerates Wolffian duct – no testosterone, anti-Mullerian hormone (AMH) not produced, and Mullerian duct can develop in addition to female reproductive organs (ovaries, vagina) XY degenerates Mullerian duct – testosterone, AMH produced, Wolffian duct continues as male reproductive organs (testes, penis) develop. -

The Migration of Paraxial and Lateral Plate Mesoderm Cells Emerging from the Late Primitive Streak Is Controlled by Different Wnt Signals
The Migration of Paraxial and Lateral Plate Mesoderm Cells Emerging from the Late Primitive Streak is Controlled by Different Wnt Signals The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Sweetman, Dylan, Laura Wagstaff, Oliver Cooper, Cornelis Weijer, and Andrea Münsterberg. 2008. The migration of paraxial and lateral plate mesoderm cells emerging from the late primitive streak is controlled by different Wnt signals. BMC Developmental Biology 8: 63. Published Version doi:10.1186/1471-213X-8-63 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:4774184 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA BMC Developmental Biology BioMed Central Research article Open Access The migration of paraxial and lateral plate mesoderm cells emerging from the late primitive streak is controlled by different Wnt signals Dylan Sweetman1, Laura Wagstaff2, Oliver Cooper1,3, Cornelis Weijer4 and Andrea Münsterberg*1 Address: 1School of Biological Sciences, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK, 2Biomedical Research Centre, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK, 3Centre for Neuroregeneration Research, Harvard Medical School, McLean Hospital, Belmont, MA 02478, USA and 4College of Life Sciences