Projected Multivariate Linear Models for Directional Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Contributions to Directional Statistics Based Clustering Methods
University of California Santa Barbara Contributions to Directional Statistics Based Clustering Methods A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Statistics & Applied Probability by Brian Dru Wainwright Committee in charge: Professor S. Rao Jammalamadaka, Chair Professor Alexander Petersen Professor John Hsu September 2019 The Dissertation of Brian Dru Wainwright is approved. Professor Alexander Petersen Professor John Hsu Professor S. Rao Jammalamadaka, Committee Chair June 2019 Contributions to Directional Statistics Based Clustering Methods Copyright © 2019 by Brian Dru Wainwright iii Dedicated to my loving wife, Carmen Rhodes, without whom none of this would have been possible, and to my sons, Max, Gus, and Judah, who have yet to know a time when their dad didn’t need to work from early till late. And finally, to my mother, Judith Moyer, without your tireless love and support from the beginning, I quite literally wouldn’t be here today. iv Acknowledgements I would like to offer my humble and grateful acknowledgement to all of the wonderful col- laborators I have had the fortune to work with during my graduate education. Much of the impetus for the ideas presented in this dissertation were derived from our work together. In particular, I would like to thank Professor György Terdik, University of Debrecen, Faculty of Informatics, Department of Information Technology. I would also like to thank Professor Saumyadipta Pyne, School of Public Health, University of Pittsburgh, and Mr. Hasnat Ali, L.V. Prasad Eye Institute, Hyderabad, India. I would like to extend a special thank you to Dr Alexander Petersen, who has held the dual role of serving on my Doctoral supervisory commit- tee as well as wearing the hat of collaborator. -

Recent Advances in Directional Statistics
Recent advances in directional statistics Arthur Pewsey1;3 and Eduardo García-Portugués2 Abstract Mainstream statistical methodology is generally applicable to data observed in Euclidean space. There are, however, numerous contexts of considerable scientific interest in which the natural supports for the data under consideration are Riemannian manifolds like the unit circle, torus, sphere and their extensions. Typically, such data can be represented using one or more directions, and directional statistics is the branch of statistics that deals with their analysis. In this paper we provide a review of the many recent developments in the field since the publication of Mardia and Jupp (1999), still the most comprehensive text on directional statistics. Many of those developments have been stimulated by interesting applications in fields as diverse as astronomy, medicine, genetics, neurology, aeronautics, acoustics, image analysis, text mining, environmetrics, and machine learning. We begin by considering developments for the exploratory analysis of directional data before progressing to distributional models, general approaches to inference, hypothesis testing, regression, nonparametric curve estimation, methods for dimension reduction, classification and clustering, and the modelling of time series, spatial and spatio- temporal data. An overview of currently available software for analysing directional data is also provided, and potential future developments discussed. Keywords: Classification; Clustering; Dimension reduction; Distributional -

A General Approach for Obtaining Wrapped Circular Distributions Via Mixtures
UC Santa Barbara UC Santa Barbara Previously Published Works Title A General Approach for obtaining wrapped circular distributions via mixtures Permalink https://escholarship.org/uc/item/5r7521z5 Journal Sankhya, 79 Author Jammalamadaka, Sreenivasa Rao Publication Date 2017 Peer reviewed eScholarship.org Powered by the California Digital Library University of California 1 23 Your article is protected by copyright and all rights are held exclusively by Indian Statistical Institute. This e-offprint is for personal use only and shall not be self- archived in electronic repositories. If you wish to self-archive your article, please use the accepted manuscript version for posting on your own website. You may further deposit the accepted manuscript version in any repository, provided it is only made publicly available 12 months after official publication or later and provided acknowledgement is given to the original source of publication and a link is inserted to the published article on Springer's website. The link must be accompanied by the following text: "The final publication is available at link.springer.com”. 1 23 Author's personal copy Sankhy¯a:TheIndianJournalofStatistics 2017, Volume 79-A, Part 1, pp. 133-157 c 2017, Indian Statistical Institute ! AGeneralApproachforObtainingWrappedCircular Distributions via Mixtures S. Rao Jammalamadaka University of California, Santa Barbara, USA Tomasz J. Kozubowski University of Nevada, Reno, USA Abstract We show that the operations of mixing and wrapping linear distributions around a unit circle commute, and can produce a wide variety of circular models. In particular, we show that many wrapped circular models studied in the literature can be obtained as scale mixtures of just the wrapped Gaussian and the wrapped exponential distributions, and inherit many properties from these two basic models. -

Bivariate Angular Estimation Under Consideration of Dependencies Using Directional Statistics
Bivariate Angular Estimation Under Consideration of Dependencies Using Directional Statistics Gerhard Kurz1, Igor Gilitschenski1, Maxim Dolgov1 and Uwe D. Hanebeck1 Abstract— Estimation of angular quantities is a widespread issue, but standard approaches neglect the true topology of the problem and approximate directional with linear uncertainties. In recent years, novel approaches based on directional statistics have been proposed. However, these approaches have been unable to consider arbitrary circular correlations between multiple angles so far. For this reason, we propose a novel recursive filtering scheme that is capable of estimating multiple angles even if they are dependent, while correctly describing their circular correlation. The proposed approach is based on toroidal probability distributions and a circular correlation coefficient. We demonstrate the superiority to a standard approach based on the Kalman filter in simulations. Fig. 1: A bivariate wrapped normal probability distribution Index Terms— recursive filtering, wrapped normal, circular on the torus shown as a heat map. correlation coefficient, moment matching. I. INTRODUCTION our knowledge, this is the first work on recursive estimation There are many applications that require estimation of based on toroidal probability distributions. angular quantities. These applications include, but are not It should be noted that there are some directional filters that, limited to, robotics, augmented reality, and aviation, as well in a sense, take correlation between angles into account. We as biology, geology, and medicine. In many cases, not just have performed research on a filter based on the hyperspheri- one, but several angles have to be estimated. Furthermore, cal Bingham distribution [8], [9], which captures correlations correlations may exist between those angles and have to be when estimating rotations represented as quaternions. -

Package 'Directional'
Package ‘Directional’ May 26, 2021 Type Package Title A Collection of R Functions for Directional Data Analysis Version 5.0 URL Date 2021-05-26 Author Michail Tsagris, Giorgos Athineou, Anamul Sajib, Eli Amson, Micah J. Waldstein Maintainer Michail Tsagris <[email protected]> Description A collection of functions for directional data (including massive data, with millions of ob- servations) analysis. Hypothesis testing, discriminant and regression analysis, MLE of distribu- tions and more are included. The standard textbook for such data is the ``Directional Statis- tics'' by Mardia, K. V. and Jupp, P. E. (2000). Other references include a) Phillip J. Paine, Si- mon P. Preston Michail Tsagris and Andrew T. A. Wood (2018). An elliptically symmetric angu- lar Gaussian distribution. Statistics and Computing 28(3): 689-697. <doi:10.1007/s11222-017- 9756-4>. b) Tsagris M. and Alenazi A. (2019). Comparison of discriminant analysis meth- ods on the sphere. Communications in Statistics: Case Studies, Data Analysis and Applica- tions 5(4):467--491. <doi:10.1080/23737484.2019.1684854>. c) P. J. Paine, S. P. Pre- ston, M. Tsagris and Andrew T. A. Wood (2020). Spherical regression models with general co- variates and anisotropic errors. Statistics and Computing 30(1): 153--165. <doi:10.1007/s11222- 019-09872-2>. License GPL-2 Imports bigstatsr, parallel, doParallel, foreach, RANN, Rfast, Rfast2, rgl RoxygenNote 6.1.1 NeedsCompilation no Repository CRAN Date/Publication 2021-05-26 15:20:02 UTC R topics documented: Directional-package . .4 A test for testing the equality of the concentration parameters for ciruclar data . .5 1 2 R topics documented: Angular central Gaussian random values simulation . -

Detecting the Direction of a Signal on High-Dimensional Spheres: Non-Null
Noname manuscript No. (will be inserted by the editor) Detecting the direction of a signal on high-dimensional spheres Non-null and Le Cam optimality results Davy Paindaveine · Thomas Verdebout Received: date / Accepted: date Abstract We consider one of the most important problems in directional statistics, namely the problem of testing the null hypothesis that the spike direction θ of a Fisher{von Mises{Langevin distribution on the p-dimensional unit hypersphere is equal to a given direction θ0. After a reduction through invariance arguments, we derive local asymptotic normality (LAN) results in a general high-dimensional framework where the dimension pn goes to infinity at an arbitrary rate with the sample size n, and where the concentration κn behaves in a completely free way with n, which offers a spectrum of problems ranging from arbitrarily easy to arbitrarily challenging ones. We identify vari- ous asymptotic regimes, depending on the convergence/divergence properties of (κn), that yield different contiguity rates and different limiting experiments. In each regime, we derive Le Cam optimal tests under specified κn and we com- pute, from the Le Cam third lemma, asymptotic powers of the classical Watson test under contiguous alternatives. We further establish LAN results with re- spect to both spike direction and concentration, which allows us to discuss optimality also under unspecified κn. To investigate the non-null behavior of the Watson test outside the parametric framework above, we derive its local Research is supported by the Program of Concerted Research Actions (ARC) of the Univer- sit´elibre de Bruxelles, by a research fellowship from the Francqui Foundation, and by the Cr´editde Recherche J.0134.18 of the FNRS (Fonds National pour la Recherche Scientifique), Communaut´eFran¸caisede Belgique. -

A General Setting for Symmetric Distributions and Their Relationship to General Distributions
Accepted Manuscript A general setting for symmetric distributions and their relationship to general distributions P.E. Jupp, G. Regoli, A. Azzalini PII: S0047-259X(16)00033-6 DOI: http://dx.doi.org/10.1016/j.jmva.2016.02.011 Reference: YJMVA 4090 To appear in: Journal of Multivariate Analysis Received date: 30 May 2015 Please cite this article as: P.E. Jupp, G. Regoli, A. Azzalini, A general setting for symmetric distributions and their relationship to general distributions, Journal of Multivariate Analysis (2016), http://dx.doi.org/10.1016/j.jmva.2016.02.011 This is a PDF file of an unedited manuscript that has been accepted for publication. Asa service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. *Manuscript Click here to download Manuscript: asymmetryR3_120216(red).pdf Click here to view linked References A general setting for symmetric distributions and their relationship to general distributions a, b c P.E. Jupp ∗, G. Regoli , A. Azzalini aSchool of Mathematics and Statistics, University of St Andrews, St Andrews KY16 9SS, UK bDipartimento di Matematica e Informatica, Universit`adi Perugia, 06123 Perugia, Italy cDipartimento di Scienze Statistiche, Universit`adi Padova, 35121 Padova, Italy Abstract A standard method of obtaining non-symmetrical distributions is that of modulating symmetrical distributions by multiplying the densities by a per- turbation factor. -

Directional Statistics: Introduction
Directional Statistics: Introduction Directional Statistics: Introduction By Riccardo Gattoa and S. Rao Jammalamadakab Keywords: Directional distributions, goodness-of-fit tests, nonparametric inference, robust estimators, rotational invariance, spacings, spacing-frequencies, two-sample tests, von Mises–Fisher distribution Abstract: In many of the natural and physical sciences, measurements are directions, either in two or three dimensions. The analysis of directional data relies on specific statistical models and procedures, which differ from the usual models and methodologies of Cartesian data. This article briefly introduces statistical models and inference for this type of data. The basic von Mises–Fisher distribution is introduced and nonparametric methods such as goodness-of-fit tests are presented. Further references are given for exploring related topics. 1 Introduction This article explores the novel area of directional statistics. In many diverse scientific fields, measurements are directions. For instance, a biologist may be measuring the direction of flight of a bird or the orientation of an animal, whereas a geologist may be interested in the direction of the Earth’s magnetic pole. Such directions may be in two dimensions as in the first two examples or in three dimensions like the last one. A set of such observations on directions is referred to as directional data (see Directional Data Analysis). Langevin[1] and Lévy[2] are two pioneering contributions in this area. One of the first statistical analyses of directional data is Fisher[3]. A two-dimensional direction is a point in ℝ2 without magnitude, for example, a unit vector. It can also be represented as a point on the circumference of the unit circle centered at the origin, denoted S1,oras an angle measured with respect to some suitably chosen “zero direction,” that is, starting point, and “sense of rotation,” that is, whether clockwise or counterclockwise is the positive direction. -

Applied Statistics Using SPSS, STATISTICA, MATLAB and R Joaquim P
Applied Statistics Using SPSS, STATISTICA, MATLAB and R Joaquim P. Marques de Sá Applied Statistics Using SPSS, STATISTICA, MATLAB and R With 195 Figures and a CD 123 E d itors Prof. Dr. Joaquim P. Marques de Sá Universidade do Porto Fac. Engenharia Rua Dr. Roberto Frias s/n 4200-465 Porto Portugal e-mail: [email protected] Library of Congress Control Number: 2007926024 ISBN 978-3-540-71971-7 Springer Berlin Heidelberg New York This work is subject to copyright. All rights are reserved, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilm or in any other way, and storage in data banks. Duplication of this publication or parts thereof is permitted only under the provisions of the German Copyright Law of September 9, 1965, in its current version, and permission for use must always be obtained from Springer. Violations are liable for prosecution under the German Copyright Law. Springer is a part of Springer Science+Business Media springer.com © Springer-Verlag Berlin Heidelberg 2007 The use of general descriptive names, registered names, trademarks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant pro- tective laws and regulations and therefore free for general use. Typesetting: by the editors Production: Integra Software Services Pvt. Ltd., India Cover design: WMX design, Heidelberg Printed on acid-free paper SPIN: 11908944 42/3100/Integra 5 4 3 2 1 0 To Wiesje and Carlos. -

The Multivariate Generalised Von Mises Distribution: Inference and Applications
Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17) The Multivariate Generalised von Mises Distribution: Inference and Applications Alexandre K. W. Navarro Jes Frellsen Richard E. Turner Department of Engineering Department of Engineering Department of Engineering University of Cambridge University of Cambridge University of Cambridge Cambridge, UK Cambridge, UK Cambridge, UK [email protected] [email protected] [email protected] Abstract Such an approach would, however, ignore the topology of the space e.g. that φ =0and φ =2π are equivalent. Alter- Circular variables arise in a multitude of data-modelling con- natively, the circular variable can be represented as a unit texts ranging from robotics to the social sciences, but they have vector in R2, x = [cos(φ), sin(φ)], and a standard bivariate been largely overlooked by the machine learning community. This paper partially redresses this imbalance by extending distribution used instead. This partially alleviates the afore- some standard probabilistic modelling tools to the circular mentioned topological problem, but standard distributions domain. First we introduce a new multivariate distribution place probability mass off the unit circle which adversely over circular variables, called the multivariate Generalised affects learning, prediction and analysis. von Mises (mGvM) distribution. This distribution can be con- In order to predict and analyse circular data it is therefore structed by restricting and renormalising a general multivari- key that machine learning practitioners have at their disposal ate Gaussian distribution to the unit hyper-torus. Previously a suite of bespoke modelling, inference and learning meth- proposed multivariate circular distributions are shown to be ods that are specifically designed for circular data (Lebanon special cases of this construction. -
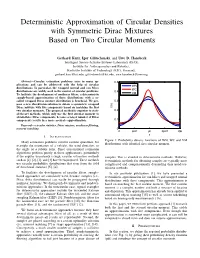
Deterministic Approximation of Circular Densities with Symmetric Dirac Mixtures Based on Two Circular Moments
Deterministic Approximation of Circular Densities with Symmetric Dirac Mixtures Based on Two Circular Moments Gerhard Kurz, Igor Gilitschenski, and Uwe D. Hanebeck Intelligent Sensor-Actuator-Systems Laboratory (ISAS), Institute for Anthropomatics and Robotics, Karlsruhe Institute of Technology (KIT), Germany, [email protected], [email protected], [email protected] Abstract—Circular estimation problems arise in many ap- 0.5 plications and can be addressed with the help of circular WN distributions. In particular, the wrapped normal and von Mises WC distributions are widely used in the context of circular problems. 0.4 To facilitate the development of nonlinear filters, a deterministic VM sample-based approximation of these distributions with a so- called wrapped Dirac mixture distribution is beneficial. We pro- 0.3 pose a new closed-form solution to obtain a symmetric wrapped Dirac mixture with five components based on matching the first f(x) two circular moments. The proposed method is superior to state- 0.2 of-the-art methods, which only use the first circular moment to obtain three Dirac components, because a larger number of Dirac components results in a more accurate approximation. 0.1 Keywords—circular statistics, Dirac mixture, nonlinear filtering, moment matching 0 0 pi/2 pi 3pi/2 2pi x I. INTRODUCTION Figure 1: Probability density functions of WN, WC and VM Many estimation problems involve circular quantities, for distributions with identical first circular moment. example the orientation of a vehicle, the wind direction, or the angle of a robotic joint. Since conventional estimation algorithms perform poorly in these applications, particularly if the angular uncertainty is high, circular estimation methods samples. -

The Power Spherical Distribution
The Power Spherical distribution Nicola De Cao 1 2 Wilker Aziz 1 Abstract = 10 0.6 p( = /2, = 4) = 100 There is a growing interest in probabilistic models 1k samples = 500 0.5 defined in hyper-spherical spaces, be it to accom- 1 modate observed data or latent structure. The von 0.4 Mises-Fisher (vMF) distribution, often regarded 0.3 0 as the Normal distribution on the hyper-sphere, is 0.2 a standard modeling choice: it is an exponential 1 0.1 1 1 family and thus enjoys important statistical results, 0 0 0.0 for example, known Kullback-Leibler (KL) diver- 0 /2 3 /2 1 1 1 2 gence from other vMF distributions. Sampling (a) On the circle S . (b) On the sphere S . from a vMF distribution, however, requires a re- jection sampling procedure which besides being Figure 1. Example of draws and density function of the Power slow poses difficulties in the context of stochas- Spherical distribution. For the circle (a) we plot both the density tic backpropagation via the reparameterization and histograms for 1k samples. For the sphere (b) we plot draws trick. Moreover, this procedure is numerically from 3 distributions of orthogonal directions (µ) and different unstable for certain vMFs, e.g., those with high concentration parameter κ. concentration and/or in high dimensions. We pro- pose a novel distribution, the Power Spherical distribution, which retains some of the important Bijral et al., 2007), mixed-membership models (Reisinger aspects of the vMF (e.g., support on the hyper- et al., 2010), computer vision (Liu et al., 2017), and natural sphere, symmetry about its mean direction param- language processing (Kumar & Tsvetkov, 2019).