Package 'Directional'
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Introduction
TR:UPM-ETSINF/DIA/2015-1 1 Univariate and bivariate truncated von Mises distributions Pablo Fernandez-Gonzalez, Concha Bielza, Pedro Larra~naga Department of Artificial Intelligence, Universidad Polit´ecnica de Madrid Abstract: In this article we study the univariate and bivariate truncated von Mises distribution, as a generalization of the von Mises distribution (Jupp and Mardia (1989)), (Mardia and Jupp (2000)). This implies the addition of two or four new truncation parameters in the univariate and, bivariate cases, respectively. The re- sults include the definition, properties of the distribution and maximum likelihood estimators for the univariate and bivariate cases. Additionally, the analysis of the bivariate case shows how the conditional distribution is a truncated von Mises dis- tribution, whereas the marginal distribution that generalizes the distribution intro- duced in Singh (2002). From the viewpoint of applications, we test the distribution with simulated data, as well as with data regarding leaf inclination angles (Bowyer and Danson. (2005)) and dihedral angles in protein chains (Murzin AG (1995)). This research aims to assert this probability distribution as a potential option for modelling or simulating any kind of phenomena where circular distributions are applicable. Key words and phrases: Angular probability distributions, Directional statistics, von Mises distribution, Truncated probability distributions. 1 Introduction The von Mises distribution has received undisputed attention in the field of di- rectional statistics (Jupp and Mardia (1989)) and in other areas like supervised classification (Lopez-Cruz et al. (2013)). Thanks to desirable properties such as its symmetry, mathematical tractability and convergence to the wrapped nor- mal distribution (Mardia and Jupp (2000)) for high concentrations, it is a viable option for many statistical analyses. -

Package 'Rfast'
Package ‘Rfast’ May 18, 2021 Type Package Title A Collection of Efficient and Extremely Fast R Functions Version 2.0.3 Date 2021-05-17 Author Manos Papadakis, Michail Tsagris, Marios Dimitriadis, Stefanos Fafalios, Ioannis Tsamardi- nos, Matteo Fasiolo, Giorgos Borboudakis, John Burkardt, Changliang Zou, Kleanthi Lakio- taki and Christina Chatzipantsiou. Maintainer Manos Papadakis <[email protected]> Depends R (>= 3.5.0), Rcpp (>= 0.12.3), RcppZiggurat LinkingTo Rcpp (>= 0.12.3), RcppArmadillo SystemRequirements C++11 BugReports https://github.com/RfastOfficial/Rfast/issues URL https://github.com/RfastOfficial/Rfast Description A collection of fast (utility) functions for data analysis. Column- and row- wise means, medians, variances, minimums, maximums, many t, F and G-square tests, many re- gressions (normal, logistic, Poisson), are some of the many fast functions. Refer- ences: a) Tsagris M., Papadakis M. (2018). Taking R to its lim- its: 70+ tips. PeerJ Preprints 6:e26605v1 <doi:10.7287/peerj.preprints.26605v1>. b) Tsagris M. and Pa- padakis M. (2018). Forward regression in R: from the extreme slow to the extreme fast. Jour- nal of Data Science, 16(4): 771--780. <doi:10.6339/JDS.201810_16(4).00006>. License GPL (>= 2.0) NeedsCompilation yes Repository CRAN Date/Publication 2021-05-17 23:50:05 UTC R topics documented: Rfast-package . .6 All k possible combinations from n elements . 10 Analysis of covariance . 11 1 2 R topics documented: Analysis of variance with a count variable . 12 Angular central Gaussian random values simulation . 13 ANOVA for two quasi Poisson regression models . 14 Apply method to Positive and Negative number . -
![Arxiv:1911.00962V1 [Cs.CV] 3 Nov 2019 Though the Label of Pose Itself Is Discrete](https://docslib.b-cdn.net/cover/7665/arxiv-1911-00962v1-cs-cv-3-nov-2019-though-the-label-of-pose-itself-is-discrete-507665.webp)
Arxiv:1911.00962V1 [Cs.CV] 3 Nov 2019 Though the Label of Pose Itself Is Discrete
Conservative Wasserstein Training for Pose Estimation Xiaofeng Liu1;2†∗, Yang Zou1y, Tong Che3y, Peng Ding4, Ping Jia4, Jane You5, B.V.K. Vijaya Kumar1 1Carnegie Mellon University; 2Harvard University; 3MILA 4CIOMP, Chinese Academy of Sciences; 5The Hong Kong Polytechnic University yContribute equally ∗Corresponding to: [email protected] Pose with true label Probability of Softmax prediction Abstract Expected probability (i.e., 1 in one-hot case) σ푁−1 푡 푠 푡푗∗ of each pose ( 푖=0 푠푖 = 1) 푗∗ 3 푠 푠 2 푠3 2 푠푗∗ 푠푗∗ This paper targets the task with discrete and periodic 푠1 푠1 class labels (e:g:; pose/orientation estimation) in the con- Same Cross-entropy text of deep learning. The commonly used cross-entropy or 푠0 푠0 regression loss is not well matched to this problem as they 푠푁−1 ignore the periodic nature of the labels and the class simi- More ideal Inferior 푠푁−1 larity, or assume labels are continuous value. We propose to Distribution 푠푁−2 Distribution 푠푁−2 incorporate inter-class correlations in a Wasserstein train- Figure 1. The limitation of CE loss for pose estimation. The ing framework by pre-defining (i:e:; using arc length of a ground truth direction of the car is tj ∗. Two possible softmax circle) or adaptively learning the ground metric. We extend predictions (green bar) of the pose estimator have the same proba- the ground metric as a linear, convex or concave increasing bility at tj ∗ position. Therefore, both predicted distributions have function w:r:t: arc length from an optimization perspective. the same CE loss. -

Contributions to Directional Statistics Based Clustering Methods
University of California Santa Barbara Contributions to Directional Statistics Based Clustering Methods A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Statistics & Applied Probability by Brian Dru Wainwright Committee in charge: Professor S. Rao Jammalamadaka, Chair Professor Alexander Petersen Professor John Hsu September 2019 The Dissertation of Brian Dru Wainwright is approved. Professor Alexander Petersen Professor John Hsu Professor S. Rao Jammalamadaka, Committee Chair June 2019 Contributions to Directional Statistics Based Clustering Methods Copyright © 2019 by Brian Dru Wainwright iii Dedicated to my loving wife, Carmen Rhodes, without whom none of this would have been possible, and to my sons, Max, Gus, and Judah, who have yet to know a time when their dad didn’t need to work from early till late. And finally, to my mother, Judith Moyer, without your tireless love and support from the beginning, I quite literally wouldn’t be here today. iv Acknowledgements I would like to offer my humble and grateful acknowledgement to all of the wonderful col- laborators I have had the fortune to work with during my graduate education. Much of the impetus for the ideas presented in this dissertation were derived from our work together. In particular, I would like to thank Professor György Terdik, University of Debrecen, Faculty of Informatics, Department of Information Technology. I would also like to thank Professor Saumyadipta Pyne, School of Public Health, University of Pittsburgh, and Mr. Hasnat Ali, L.V. Prasad Eye Institute, Hyderabad, India. I would like to extend a special thank you to Dr Alexander Petersen, who has held the dual role of serving on my Doctoral supervisory commit- tee as well as wearing the hat of collaborator. -

A Guide on Probability Distributions
powered project A guide on probability distributions R-forge distributions Core Team University Year 2008-2009 LATEXpowered Mac OS' TeXShop edited Contents Introduction 4 I Discrete distributions 6 1 Classic discrete distribution 7 2 Not so-common discrete distribution 27 II Continuous distributions 34 3 Finite support distribution 35 4 The Gaussian family 47 5 Exponential distribution and its extensions 56 6 Chi-squared's ditribution and related extensions 75 7 Student and related distributions 84 8 Pareto family 88 9 Logistic ditribution and related extensions 108 10 Extrem Value Theory distributions 111 3 4 CONTENTS III Multivariate and generalized distributions 116 11 Generalization of common distributions 117 12 Multivariate distributions 132 13 Misc 134 Conclusion 135 Bibliography 135 A Mathematical tools 138 Introduction This guide is intended to provide a quite exhaustive (at least as I can) view on probability distri- butions. It is constructed in chapters of distribution family with a section for each distribution. Each section focuses on the tryptic: definition - estimation - application. Ultimate bibles for probability distributions are Wimmer & Altmann (1999) which lists 750 univariate discrete distributions and Johnson et al. (1994) which details continuous distributions. In the appendix, we recall the basics of probability distributions as well as \common" mathe- matical functions, cf. section A.2. And for all distribution, we use the following notations • X a random variable following a given distribution, • x a realization of this random variable, • f the density function (if it exists), • F the (cumulative) distribution function, • P (X = k) the mass probability function in k, • M the moment generating function (if it exists), • G the probability generating function (if it exists), • φ the characteristic function (if it exists), Finally all graphics are done the open source statistical software R and its numerous packages available on the Comprehensive R Archive Network (CRAN∗). -

Recent Advances in Directional Statistics
Recent advances in directional statistics Arthur Pewsey1;3 and Eduardo García-Portugués2 Abstract Mainstream statistical methodology is generally applicable to data observed in Euclidean space. There are, however, numerous contexts of considerable scientific interest in which the natural supports for the data under consideration are Riemannian manifolds like the unit circle, torus, sphere and their extensions. Typically, such data can be represented using one or more directions, and directional statistics is the branch of statistics that deals with their analysis. In this paper we provide a review of the many recent developments in the field since the publication of Mardia and Jupp (1999), still the most comprehensive text on directional statistics. Many of those developments have been stimulated by interesting applications in fields as diverse as astronomy, medicine, genetics, neurology, aeronautics, acoustics, image analysis, text mining, environmetrics, and machine learning. We begin by considering developments for the exploratory analysis of directional data before progressing to distributional models, general approaches to inference, hypothesis testing, regression, nonparametric curve estimation, methods for dimension reduction, classification and clustering, and the modelling of time series, spatial and spatio- temporal data. An overview of currently available software for analysing directional data is also provided, and potential future developments discussed. Keywords: Classification; Clustering; Dimension reduction; Distributional -

Chapter 2 Parameter Estimation for Wrapped Distributions
ABSTRACT RAVINDRAN, PALANIKUMAR. Bayesian Analysis of Circular Data Using Wrapped Dis- tributions. (Under the direction of Associate Professor Sujit K. Ghosh). Circular data arise in a number of different areas such as geological, meteorolog- ical, biological and industrial sciences. We cannot use standard statistical techniques to model circular data, due to the circular geometry of the sample space. One of the com- mon methods used to analyze such data is the wrapping approach. Using the wrapping approach, we assume that, by wrapping a probability distribution from the real line onto the circle, we obtain the probability distribution for circular data. This approach creates a vast class of probability distributions that are flexible to account for different features of circular data. However, the likelihood-based inference for such distributions can be very complicated and computationally intensive. The EM algorithm used to compute the MLE is feasible, but is computationally unsatisfactory. Instead, we use Markov Chain Monte Carlo (MCMC) methods with a data augmentation step, to overcome such computational difficul- ties. Given a probability distribution on the circle, we assume that the original distribution was distributed on the real line, and then wrapped onto the circle. If we can unwrap the distribution off the circle and obtain a distribution on the real line, then the standard sta- tistical techniques for data on the real line can be used. Our proposed methods are flexible and computationally efficient to fit a wide class of wrapped distributions. Furthermore, we can easily compute the usual summary statistics. We present extensive simulation studies to validate the performance of our method. -

A Compendium of Conjugate Priors
A Compendium of Conjugate Priors Daniel Fink Environmental Statistics Group Department of Biology Montana State Univeristy Bozeman, MT 59717 May 1997 Abstract This report reviews conjugate priors and priors closed under sampling for a variety of data generating processes where the prior distributions are univariate, bivariate, and multivariate. The effects of transformations on conjugate prior relationships are considered and cases where conjugate prior relationships can be applied under transformations are identified. Univariate and bivariate prior relationships are verified using Monte Carlo methods. Contents 1 Introduction Experimenters are often in the position of having had collected some data from which they desire to make inferences about the process that produced that data. Bayes' theorem provides an appealing approach to solving such inference problems. Bayes theorem, π(θ) L(θ x ; : : : ; x ) g(θ x ; : : : ; x ) = j 1 n (1) j 1 n π(θ) L(θ x ; : : : ; x )dθ j 1 n is commonly interpreted in the following wayR. We want to make some sort of inference on the unknown parameter(s), θ, based on our prior knowledge of θ and the data collected, x1; : : : ; xn . Our prior knowledge is encapsulated by the probability distribution on θ; π(θ). The data that has been collected is combined with our prior through the likelihood function, L(θ x ; : : : ; x ) . The j 1 n normalized product of these two components yields a probability distribution of θ conditional on the data. This distribution, g(θ x ; : : : ; x ) , is known as the posterior distribution of θ. Bayes' j 1 n theorem is easily extended to cases where is θ multivariate, a vector of parameters. -

A General Approach for Obtaining Wrapped Circular Distributions Via Mixtures
UC Santa Barbara UC Santa Barbara Previously Published Works Title A General Approach for obtaining wrapped circular distributions via mixtures Permalink https://escholarship.org/uc/item/5r7521z5 Journal Sankhya, 79 Author Jammalamadaka, Sreenivasa Rao Publication Date 2017 Peer reviewed eScholarship.org Powered by the California Digital Library University of California 1 23 Your article is protected by copyright and all rights are held exclusively by Indian Statistical Institute. This e-offprint is for personal use only and shall not be self- archived in electronic repositories. If you wish to self-archive your article, please use the accepted manuscript version for posting on your own website. You may further deposit the accepted manuscript version in any repository, provided it is only made publicly available 12 months after official publication or later and provided acknowledgement is given to the original source of publication and a link is inserted to the published article on Springer's website. The link must be accompanied by the following text: "The final publication is available at link.springer.com”. 1 23 Author's personal copy Sankhy¯a:TheIndianJournalofStatistics 2017, Volume 79-A, Part 1, pp. 133-157 c 2017, Indian Statistical Institute ! AGeneralApproachforObtainingWrappedCircular Distributions via Mixtures S. Rao Jammalamadaka University of California, Santa Barbara, USA Tomasz J. Kozubowski University of Nevada, Reno, USA Abstract We show that the operations of mixing and wrapping linear distributions around a unit circle commute, and can produce a wide variety of circular models. In particular, we show that many wrapped circular models studied in the literature can be obtained as scale mixtures of just the wrapped Gaussian and the wrapped exponential distributions, and inherit many properties from these two basic models. -

Recursive Bayesian Filtering in Circular State Spaces
Recursive Bayesian Filtering in Circular State Spaces Gerhard Kurza, Igor Gilitschenskia, Uwe D. Hanebecka aIntelligent Sensor-Actuator-Systems Laboratory (ISAS) Institute for Anthropomatics and Robotics Karlsruhe Institute of Technology (KIT), Germany Abstract For recursive circular filtering based on circular statistics, we introduce a general framework for estimation of a circular state based on different circular distributions, specifically the wrapped normal distribution and the von Mises distribution. We propose an estimation method for circular systems with nonlinear system and measurement functions. This is achieved by relying on efficient deterministic sampling techniques. Furthermore, we show how the calculations can be simplified in a variety of important special cases, such as systems with additive noise as well as identity system or measurement functions. We introduce several novel key components, particularly a distribution-free prediction algorithm, a new and superior formula for the multiplication of wrapped normal densities, and the ability to deal with non-additive system noise. All proposed methods are thoroughly evaluated and compared to several state-of-the-art solutions. 1. Introduction Estimation of circular quantities is an omnipresent issue, be it the wind direction, the angle of a robotic revolute joint, the orientation of a turntable, or the direction a vehicle is facing. Circular estimation is not limited to applications involving angles, however, and can be applied to a variety of periodic phenomena. For example phase estimation is a common issue in signal processing, and tracking objects that periodically move along a certain trajectory is also of interest. Standard approaches to circular estimation are typically based on estimation techniques designed for linear scenarios that are tweaked to deal with some of the issues arising in the presence of circular quantities. -
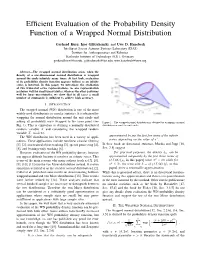
Efficient Evaluation of the Probability Density Function
Efficient Evaluation of the Probability Density Function of a Wrapped Normal Distribution Gerhard Kurz, Igor Gilitschenski, and Uwe D. Hanebeck Intelligent Sensor-Actuator-Systems Laboratory (ISAS) Institute for Anthropomatics and Robotics Karlsruhe Institute of Technology (KIT), Germany [email protected], [email protected], [email protected] Abstract—The wrapped normal distribution arises when the density of a one-dimensional normal distribution is wrapped around the circle infinitely many times. At first look, evaluation of its probability density function appears tedious as an infinite series is involved. In this paper, we investigate the evaluation of two truncated series representations. As one representation performs well for small uncertainties, whereas the other performs well for large uncertainties, we show that in all cases a small number of summands is sufficient to achieve high accuracy. I. INTRODUCTION The wrapped normal (WN) distribution is one of the most widely used distributions in circular statistics. It is obtained by wrapping the normal distribution around the unit circle and adding all probability mass wrapped to the same point (see Figure 1. The wrapped normal distribution is obtained by wrapping a normal Fig. 1). This is equivalent to defining a normally distributed distribution around the unit circle. random variable X and considering the wrapped random variable X mod 2π. approximated by just the first few terms of the infinite The WN distribution has been used in a variety of appli- series, depending on the value of σ2. cations. These applications include nonlinear circular filtering [1], [2], constrained object tracking [3], speech processing [4], In their book on directional statistics, Mardia and Jupp [16, [5], and bearings-only tracking [6]. -

Multivariate Statistical Functions in R
Multivariate statistical functions in R Michail T. Tsagris [email protected] College of engineering and technology, American university of the middle east, Egaila, Kuwait Version 6.1 Athens, Nottingham and Abu Halifa (Kuwait) 31 October 2014 Contents 1 Mean vectors 1 1.1 Hotelling’s one-sample T2 test ............................. 1 1.2 Hotelling’s two-sample T2 test ............................ 2 1.3 Two two-sample tests without assuming equality of the covariance matrices . 4 1.4 MANOVA without assuming equality of the covariance matrices . 6 2 Covariance matrices 9 2.1 One sample covariance test .............................. 9 2.2 Multi-sample covariance matrices .......................... 10 2.2.1 Log-likelihood ratio test ............................ 10 2.2.2 Box’s M test ................................... 11 3 Regression, correlation and discriminant analysis 13 3.1 Correlation ........................................ 13 3.1.1 Correlation coefficient confidence intervals and hypothesis testing us- ing Fisher’s transformation .......................... 13 3.1.2 Non-parametric bootstrap hypothesis testing for a zero correlation co- efficient ..................................... 14 3.1.3 Hypothesis testing for two correlation coefficients . 15 3.2 Regression ........................................ 15 3.2.1 Classical multivariate regression ....................... 15 3.2.2 k-NN regression ................................ 17 3.2.3 Kernel regression ................................ 20 3.2.4 Choosing the bandwidth in kernel regression in a very simple way . 23 3.2.5 Principal components regression ....................... 24 3.2.6 Choosing the number of components in principal component regression 26 3.2.7 The spatial median and spatial median regression . 27 3.2.8 Multivariate ridge regression ......................... 29 3.3 Discriminant analysis .................................. 31 3.3.1 Fisher’s linear discriminant function ....................