A Cyanobacterial Photorespiratory Bypass Model to Enhance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Proteomic Analysis of Mycelial Proteins from Magnaporthe Oryzae Under Nitrogen Starvation
Proteomic analysis of mycelial proteins from Magnaporthe oryzae under nitrogen starvation X.-G. Zhou1,2, P. Yu 2, C. Dong2, C.-X. Yao2, Y.-M. Ding2, N. Tao2 and Z.-W. Zhao1 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China 2Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Yunnan Provincial Key Laboratory of Agricultural Biotechnology, Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences, Kunming, China Corresponding author: Z.W. Zhao E-mail: [email protected] Genet. Mol. Res. 15 (2): gmr.15028637 Received March 23, 2016 Accepted April 11, 2016 Published May 13, 2016 DOI http://dx.doi.org/10.4238/gmr.15028637 ABSTRACT. Magnaporthe oryzae is an important model system in studies of plant pathogenic fungi, and nitrogen is a key nutrient source affecting microbial growth and development. In order to understand how nitrogen stress causes changes in mycelial proteins, we analyzed differentially expressed mycelial proteins from the M. oryzae virulent strain CH-63 using two-dimensional electrophoresis and mass spectrometry in complete medium or under nitrogen starvation conditions. A total of 975 ± 70 and 1169 ± 90 protein spots were detected in complete medium and under nitrogen starvation conditions, respectively. Forty-nine protein spots exhibited at least 2-fold up- regulation or down-regulation at the protein level according to PDQuest7.4. Moreover, 43 protein spots were successfully identified by matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight mass spectrometry. Among these spots, 6 proteins were functionally unknown and 37 proteins were categorized into 5 groups according to Genetics and Molecular Research 15 (2): gmr.15028637 ©FUNPEC-RP www.funpecrp.com.br X.-G. -

Molecular Biology for Computer Scientists
CHAPTER 1 Molecular Biology for Computer Scientists Lawrence Hunter “Computers are to biology what mathematics is to physics.” — Harold Morowitz One of the major challenges for computer scientists who wish to work in the domain of molecular biology is becoming conversant with the daunting intri- cacies of existing biological knowledge and its extensive technical vocabu- lary. Questions about the origin, function, and structure of living systems have been pursued by nearly all cultures throughout history, and the work of the last two generations has been particularly fruitful. The knowledge of liv- ing systems resulting from this research is far too detailed and complex for any one human to comprehend. An entire scientific career can be based in the study of a single biomolecule. Nevertheless, in the following pages, I attempt to provide enough background for a computer scientist to understand much of the biology discussed in this book. This chapter provides the briefest of overviews; I can only begin to convey the depth, variety, complexity and stunning beauty of the universe of living things. Much of what follows is not about molecular biology per se. In order to 2ARTIFICIAL INTELLIGENCE & MOLECULAR BIOLOGY explain what the molecules are doing, it is often necessary to use concepts involving, for example, cells, embryological development, or evolution. Bi- ology is frustratingly holistic. Events at one level can effect and be affected by events at very different levels of scale or time. Digesting a survey of the basic background material is a prerequisite for understanding the significance of the molecular biology that is described elsewhere in the book. -

Oxdc Antibody Rabbit Polyclonal Antibody Catalog # ABV11223
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 OxdC Antibody Rabbit Polyclonal Antibody Catalog # ABV11223 Specification OxdC Antibody - Product Information Application WB Primary Accession O34714 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit IgG Calculated MW 43566 OxdC Antibody - Additional Information Gene ID 938620 Positive Control Western Blot: Recombinant protein Application & Usage Western blot: 1-4 Western blot of Oxalate decarboxylase µg/ml. antibody. Lane 1: rb- Oxalate decarboxylase - Other Names 10 ng. Lane 2: rb- Oxalate decarboxylase - 50 YvrK ng Target/Specificity OxdC OxdC Antibody - Background Antibody Form Oxalate decarboxylase (OxdC, EC4.1.1.2) is a Liquid manganese-containing enzyme, which decomposes oxalic acid and oxalate. With Appearance OxdC catalysis, oxalate is split into formate Colorless liquid and CO2. This enzyme belongs to the family of lyases, specifically the carboxy-lyases, which Formulation 100 µg (0.5 mg/ml) of antibody in PBS pH cleave carbon-carbon bonds. The systematic 7.2 containing 0.01 % BSA, 0.01 % name of this enzyme class is oxalate thimerosal, and 50 % glycerol. carboxy-lyase (formate-forming). This enzyme is also called oxalate carboxy-lyase. The Handling enzyme is composed of two cupin domains, The antibody solution should be gently each of which contains a Mn (II) ion. This mixed before use. enzyme participates in glyoxylate and dicarboxylate metabolism. This enzyme has Reconstitution & Storage been recognized for diagnostics in diverse -20 °C biotechnological applications such as the clinical assay of oxalate in blood and urine, Background Descriptions therapeutics, process industry, and agriculture to lower oxalate levels in foods and the environment. -

Department of Pharmacology (GRAD) 1
Department of Pharmacology (GRAD) 1 faculty participate fully at all levels. The department has the highest DEPARTMENT OF level of NIH funding of all pharmacology departments and a great diversity of research areas is available to trainees. These areas PHARMACOLOGY (GRAD) include cell surface receptors, G proteins, protein kinases, and signal transduction mechanisms; neuropharmacology; nucleic acids, cancer, Contact Information and antimicrobial pharmacology; and experimental therapeutics. Cell and molecular approaches are particularly strong, but systems-level research Department of Pharmacology such as behavioral pharmacology and analysis of knock-in and knock-out Visit Program Website (http://www.med.unc.edu/pharm/) mice is also well-represented. Excellent physical facilities are available for all research areas. Henrik Dohlman, Chair Students completing the training program will have acquired basic The Department of Pharmacology offers a program of study that leads knowledge of pharmacology and related fields, in-depth knowledge in to the degree of doctor of philosophy in pharmacology. The curriculum is their dissertation research area, the ability to evaluate scientific literature, individualized in recognition of the diverse backgrounds and interests of mastery of a variety of laboratory procedures, skill in planning and students and the broad scope of the discipline of pharmacology. executing an important research project in pharmacology, and the ability The department offers a variety of research areas including to communicate results, analysis, and interpretation. These skills provide a sound basis for successful scientific careers in academia, government, 1. Receptors and signal transduction or industry. 2. Ion channels To apply to BBSP, students must use The Graduate School's online 3. -

Proteo-Metabolomic Investigation of Transgenic Rice Unravels Metabolic
www.nature.com/scientificreports OPEN Proteo-metabolomic investigation of transgenic rice unravels metabolic alterations and Received: 27 November 2018 Accepted: 24 June 2019 accumulation of novel proteins Published: xx xx xxxx potentially involved in defence against Rhizoctonia solani Subhasis Karmakar1, Karabi Datta1, Kutubuddin Ali Molla2,3, Dipak Gayen4, Kaushik Das1, Sailendra Nath Sarkar1 & Swapan K. Datta1 The generation of sheath blight (ShB)-resistant transgenic rice plants through the expression of Arabidopsis NPR1 gene is a signifcant development for research in the feld of biotic stress. However, to our knowledge, regulation of the proteomic and metabolic networks in the ShB-resistant transgenic rice plants has not been studied. In the present investigation, the relative proteome and metabolome profles of the non–transformed wild-type and the AtNPR1-transgenic rice lines prior to and subsequent to the R. solani infection were investigated. Total proteins from wild type and transgenic plants were investigated using two-dimensional gel electrophoresis (2-DE) followed by mass spectrometry (MS). The metabolomics study indicated an increased abundance of various metabolites, which draws parallels with the proteomic analysis. Furthermore, the proteome data was cross-examined using network analysis which identifed modules that were rich in known as well as novel immunity-related prognostic proteins, particularly the mitogen-activated protein kinase 6, probable protein phosphatase 2C1, probable trehalose-phosphate phosphatase 2 and heat shock protein. A novel protein, 14–3– 3GF14f was observed to be upregulated in the leaves of the transgenic rice plants after ShB infection, and the possible mechanistic role of this protein in ShB resistance may be investigated further. -

Bioinorganic Chemistry Content
Bioinorganic Chemistry Content 1. What is bioinorganic chemistry? 2. Evolution of elements 3. Elements and molecules of life 4. Phylogeny 5. Metals in biochemistry 6. Ligands in biochemistry 7. Principals of coordination chemistry 8. Properties of bio molecules 9. Biochemistry of main group elements 10. Biochemistry of transition metals 11. Biochemistry of lanthanides and actinides 12. Modell complexes 13. Analytical methods in bioinorganic 14. Applications areas of bioinorganic chemistry "Simplicity is the ultimate sophistication" Leonardo Da Vinci Bioinorganic Chemistry Slide 1 Prof. Dr. Thomas Jüstel Literature • C. Elschenbroich, A. Salzer, Organometallchemie, 2. Auflage, Teubner, 1988 • S.J. Lippard, J.N. Berg, Bioinorganic Chemistry, Spektrum Akademischer Verlag, 1995 • J.E. Huheey, E. Keiter, R. Keiter, Anorganische Chemie – Prinzipien von Struktur und Reaktivität, 3. Auflage, Walter de Gruyter, 2003 • W. Kaim, B. Schwederski: Bioinorganic Chemistry, 4. Auflage, Vieweg-Teubner, 2005 • H. Rauchfuß, Chemische Evolution und der Ursprung des Lebens, Springer, 2005 • A.F. Hollemann, N. Wiberg, Lehrbuch der Anorganischen Chemie, 102. Auflage, de Gruyter, 2007 • I. Bertini, H.B. Gray, E.I. Stiefel, J.S. Valentine, Biological Chemistry, University Science Books, 2007 • N. Metzler-Nolte, U. Schatzschneier, Bioinorganic Chemistry: A Practical Course, Walter de Gruyter, 2009 • W. Ternes, Biochemie der Elemente, Springer, 2013 • D. Rabinovich, Bioinorganic Chemistry, Walter de Gruyter, 2020 Bioinorganic Chemistry Slide 2 Prof. Dr. Thomas Jüstel 1. What is Bioinorganic Chemistry? A Highly Interdisciplinary Science at the Verge of Biology, Chemistry, Physics, and Medicine Biochemistry Inorganic Chemistry (Micro)- Physics & Biology Spectroscopy Bioinorganic Chemistry Pharmacy & Medicine & Toxicology Physiology Diagnostics Bioinorganic Chemistry Slide 3 Prof. Dr. Thomas Jüstel 2. Evolution of the Elements Most Abundant Elements in the Universe According to Atomic Fraction Are: 1. -
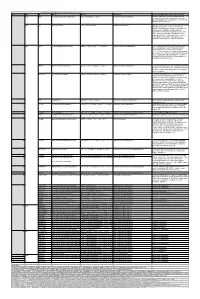
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -

Independent Research Resources Demonstrations/Simulations
Independent Research Resources Independent Generation of Research (IGoR) - IGoR provides a platform for people to pool their knowledge, resources, time, and creativity so that everyone can pursue their own scientific curiosity. Virginia Junior Academy of Science Resource Library - Extensive collection of open-access resources for students in Biology & Medicine, Botany, Ecology, Environmental Sciences, Chemistry, Engineering and Physics The Society for Science and the Public Science Project Resources - A catalog of science resources that can support your quest to learn and do science Science Buddies - Ideas for science projects Teacher resources National Center for Science Education Scientist in the Classroom - Platform allows teachers to request classroom visits from scientists Genetics Education Outreach Network (GEON) - Network of genetics professionals HHMI BioInteractive Data Points - Explore and interpret primary data from published research Biotech in a Box Loan Kits - Shipped to your school from Fralin Life Sciences Institute at Virginia Tech Demonstrations/Simulations Genetic Science Learning Center- Simulations, videos and interactive activities that explore genetics, cell biology, neuroscience, ecology and health Remotely Accessible Instruments for Nanotechnology (RAIN) - Access and control nanoinstruments over the Internet in real-time with the assistance of an experienced engineer PhET Simulations - Interactive STEM simulations for all grade levels HHMI BioInteractive Interactive Media - Recommendations: Virus Explorer; Exploring -

Oxalate Decarboxylase from Bacillus Subtilis, Recombinant Cat
Oxalate Decarboxylase from Bacillus subtilis, recombinant Cat. No. NATE-1688 Lot. No. (See product label) Introduction Description Oxalate decarboxylase (OxdC, EC4.1.1.2) is a manganese-containing enzyme, which decomposes oxalic acid and oxalate. With OxdC catalysis, oxalate is split into formate and CO2. This enzyme belongs to the family of lyases, specifically the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is oxalate carboxy-lyase (formate-forming). This enzyme is also called oxalate carboxy-lyase. The enzyme is composed of two cupin domains, each of which contains a Mn (II) ion. This enzyme participates in glyoxylate and dicarboxylate metabolism. This enzyme has been recognized for diagnostics in diverse biotechnological applications such as the clinical assay of oxalate in blood and urine, therapeutics, process industry, and agriculture to lower oxalate levels in foods and the environment. The recombinant protein made from the Bacillus Subtilis sequence includes OxdC with N-terminal His-tag. Synonyms Oxalate carboxy-lyase; EC 4.1.1.2; Oxalate decarboxylase; OxdC Product Information Species Bacillus subtilis Source E. coli Form Liquid Formulation In 50 mM NaOAC, pH 5.5. The activation was stopped by addition of 10 mM MMTS, which can be removed under reducing conditions. EC Number EC 4.1.1.2 CAS No. 9024-97-9 Molecular Weight 45.9 kDa Purity > 98% by SDS-PAGE Activity 150U/mg Creative Enzymes. All rights reserved. 45-1 Ramsey Road, Shirley, NY 11967, USA Tel:1-631-562-8517 1-516-512-3133 Fax:1-631-938-8127 E-mail: [email protected] http://www.creative-enzymes.com Concentration 2 mg/mL Unit Definition One unit is the amount of enzyme that generates 1.0 µmole of NADH at 37°C. -

The Structure of Oxalate Decarboxylase at Its Active Ph
bioRxivRunning preprint head: doi: https://doi.org/10.1101/426874 Low pH Structure, Oxalate; this version Decarboxylase posted September 25, 2018. The copyright holder for this preprint (which was1 not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Structure of Oxalate Decarboxylase at its Active pH M. J. Burg, J. L. Goodsell, U. T. Twahir, S. D. Bruner, and A. Angerhofer, bioRxivLow preprintpH Structure, doi: https://doi.org/10.1101/426874 Oxalate Decarboxylase; this version posted September 25, 2018. The copyright holder for this preprint (which was2 not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Oxalate decarboxylase catalyzes the redox-neutral unimolecular disproportionation reaction of oxalic acid. The pH maximum for catalysis is ~4.0 and activity is negligible above pH7. Here we report on the first crystal structure of the enzyme in its active pH range at pH4.6, and at a resolution of 1.45 Å, the highest to date. The fundamental tertiary and quaternary structure of the enzyme does not change with pH. However, the low pH crystals are heterogeneous containing both a closed and open conformation of a flexible loop region which gates access to the N-terminal active site cavity. Residue E162 in the closed conformation points away from the active-site Mn ion owing to the coordination of a buffer molecule, acetate. Since the quaternary structure of the enzyme appears unaffected by pH many conclusions drawn from the structures taken at high pH remain valid. -

Biochemistry and Molecular Biology
School of Life Sciences / Department of Biochemistry and Biophysics BIOCHEMISTRY AND MOLECULAR BIOLOGY Revealing how life works Department of Biochemistry and Biophysics Oregon State University 2011 Agriculture and Life Sciences Building Corvallis, OR 97331 OSUBB 541-737-4511 biochem.oregonstate.edu College of Science Oregon State University 128 Kidder Hall Corvallis, OR 97331 541-737-4811 science.oregonstate.edu This publication will be made available in an accessible alternative format upon request. Please contact the College COLLEGE OF SCIENCE of Science at 541-737-4811 or [email protected]. ACADEMIC BROCHURE / 2020 Highlights • Solve problems at the What will you discover? intersection of biological and physical sciences with our The Department of Biochemistry and Biophysics offers nationally accredited program world-class faculty, a tradition of interdisciplinary in biochemistry, molecular and research, teaching excellence and extraordinary cellular biology, chemistry, laboratories to facilitate undergraduate learning. molecular genetics, physics and statistics. The department ranks high nationally and internationally in many research areas, including signal transduction, gene • Tailor your education to expression, epigenetics, metabolic regulation, structural specific career goals with three biology, and genetic code expansion technology. different options. The department offers two Bachelor of Science degrees, • Thrive in a smaller, supportive both accredited by the American Society for Biochemistry department within a world-class and Molecular Biology (ASBMB): research university. • Biochemistry and Molecular Biology (BMB) with • Pursue interdisciplinary research options in Advanced Molecular Biology, Computational projects with faculty across OSU. Molecular Biology, and Pre-medicine; • Participate in the Biochemistry • Biochemistry and Biophysics (BB) with options in Club for community, leadership Advanced Biophysics, Neuroscience, and Pre-medicine.