Thyroid Hormone Receptor SS
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Reconstruction of the global neural crest gene regulatory network in vivo Ruth M Williams1, Ivan Candido-Ferreira1, Emmanouela Repapi2, Daria Gavriouchkina1,4, Upeka Senanayake1, Jelena Telenius2,3, Stephen Taylor2, Jim Hughes2,3, and Tatjana Sauka-Spengler1,∗ Supplemental Material ∗Lead and corresponding author: Tatjana Sauka-Spengler ([email protected]) 1University of Oxford, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, Oxford, OX3 9DS, UK 2University of Oxford, MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 3University of Oxford, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 4Present Address: Okinawa Institute of Science and Technology, Molecular Genetics Unit, Onna, 904-0495, Japan A 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R1_5-6ss) log2(R1_8-10ss) log2(R1_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.92 0 r=0.99 0 r=0.96 0 r=0.99 0 r=0.96 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R2_non-NC) log2(R2_5-6ss) log2(R3_5-6ss) log2(R2_8-10ss) log2(R3_8-10ss) 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R2_5-6ss) log2(R1_8-10ss) log2(R2_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.94 0 r=0.96 0 r=0.95 0 r=0.96 0 r=0.95 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R3_non-NC) log2(R4_5-6ss) log2(R3_5-6ss) log2(R4_8-10ss) log2(R3_8-10ss) -

A Cell Line P53 Mutation Type UM
A Cell line p53 mutation Type UM-SCC 1 wt UM-SCC5 Exon 5, 157 GTC --> TTC Missense mutation by transversion (Valine --> Phenylalanine UM-SCC6 wt UM-SCC9 wt UM-SCC11A wt UM-SCC11B Exon 7, 242 TGC --> TCC Missense mutation by transversion (Cysteine --> Serine) UM-SCC22A Exon 6, 220 TAT --> TGT Missense mutation by transition (Tyrosine --> Cysteine) UM-SCC22B Exon 6, 220 TAT --> TGT Missense mutation by transition (Tyrosine --> Cysteine) UM-SCC38 Exon 5, 132 AAG --> AAT Missense mutation by transversion (Lysine --> Asparagine) UM-SCC46 Exon 8, 278 CCT --> CGT Missense mutation by transversion (Proline --> Alanine) B 1 Supplementary Methods Cell Lines and Cell Culture A panel of ten established HNSCC cell lines from the University of Michigan series (UM-SCC) was obtained from Dr. T. E. Carey at the University of Michigan, Ann Arbor, MI. The UM-SCC cell lines were derived from eight patients with SCC of the upper aerodigestive tract (supplemental Table 1). Patient age at tumor diagnosis ranged from 37 to 72 years. The cell lines selected were obtained from patients with stage I-IV tumors, distributed among oral, pharyngeal and laryngeal sites. All the patients had aggressive disease, with early recurrence and death within two years of therapy. Cell lines established from single isolates of a patient specimen are designated by a numeric designation, and where isolates from two time points or anatomical sites were obtained, the designation includes an alphabetical suffix (i.e., "A" or "B"). The cell lines were maintained in Eagle's minimal essential media supplemented with 10% fetal bovine serum and penicillin/streptomycin. -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -
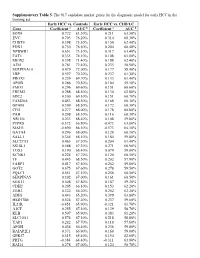
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

Structural Heterogeneity of Cellular K5/K14 Filaments As Revealed by Cryo
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.12.442145; this version posted May 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Structural heterogeneity of cellular K5/K14 filaments as revealed by cryo- 2 electron microscopy 3 4 Short title: Structural heterogeneity of keratin filaments 5 6 7 Miriam S. Weber1, Matthias Eibauer1, Suganya Sivagurunathan2, Thomas M. Magin3, Robert D. 8 Goldman2, Ohad Medalia1* 9 1Department of Biochemistry, University of Zurich, Switzerland 10 2Department of Cell and Developmental Biology, Northwestern University Feinberg School of 11 Medicine, USA 12 3Institute of Biology, University of Leipzig, Germany 13 14 * Corresponding author: [email protected] 15 16 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.05.12.442145; this version posted May 14, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 17 Abstract 18 Keratin intermediate filaments are an essential and major component of the cytoskeleton in epithelial 19 cells. They form a stable yet dynamic filamentous network extending from the nucleus to the cell 20 periphery. Keratin filaments provide cellular resistance to mechanical stresses, ensure cell and tissue 21 integrity in addition to regulatory functions. Mutations in keratin genes are related to a variety of 22 epithelial tissue diseases that mostly affect skin and hair. Despite their importance, the molecular 23 structure of keratin filaments remains largely unknown. In this study, we analyzed the structure of 24 keratin 5/keratin 14 filaments within ghost keratinocytes by cryo-electron microscopy and cryo- 25 electron tomography. -

P63 Transcription Factor Regulates Nuclear Shape and Expression of Nuclear Envelope
Durham Research Online Deposited in DRO: 21 September 2017 Version of attached le: Published Version Peer-review status of attached le: Peer-reviewed Citation for published item: Rapisarda, Valentina and Malashchuk, Igor and Asamaowei, Inemo E. and Poterlowicz, Krzysztof and Fessing, Michael Y. and Sharov, Andrey A. and Karakesisoglou, Iakowos and Botchkarev, Vladimir A. and Mardaryev, Andrei (2017) 'p63 transcription factor regulates nuclear shape and expression of nuclear envelope-associated genes in epidermal keratinocytes.', Journal of investigative dermatology., 137 (10). pp. 2157-2167. Further information on publisher's website: https://doi.org/10.1016/j.jid.2017.05.013 Publisher's copyright statement: This article is available under the terms of the Creative Commons Attribution License (CC BY). You may copy and distribute the article, create extracts, abstracts and new works from the article, alter and revise the article, text or data mine the article and otherwise reuse the article commercially (including reuse and/or resale of the article) without permission from Elsevier. You must give appropriate credit to the original work, together with a link to the formal publication through the relevant DOI and a link to the Creative Commons user license above. You must indicate if any changes are made but not in any way that suggests the licensor endorses you or your use of the work. Additional information: Use policy The full-text may be used and/or reproduced, and given to third parties in any format or medium, without prior permission or charge, for personal research or study, educational, or not-for-prot purposes provided that: • a full bibliographic reference is made to the original source • a link is made to the metadata record in DRO • the full-text is not changed in any way The full-text must not be sold in any format or medium without the formal permission of the copyright holders. -

Knock-Out Validated Antibodies from Cloud-Clone Cat.No
Knock-out validated antibodies from Cloud-Clone Cat.No. Target PAA778Hu01 B-Cell Leukemia/Lymphoma 2 (Bcl2) PAL763Hu01 Myxovirus Resistance 1 (MX1) PAB698Hu01 Lactate Dehydrogenase B (LDHB) PAA009Hu01 Angiopoietin 2 (ANGPT2) PAA849Ra01 Glycogen Phosphorylase, Liver (PYGL) PAA153Hu01 Alpha-Fetoprotein (aFP) PAF460Hu01 Folate Receptor 1, Adult (FOLR1) PAB233Hu01 Cyclin Dependent Kinase 4 (CDK4) PAA150Hu04 Carcinoembryonic Antigen (CEA) PAB905Hu01 Interleukin 7 Receptor (IL7R) PAC823Hu01 Thymidine Kinase 1, Soluble (TK1) PAH838Hu01 Isocitrate Dehydrogenase 2, mitochondrial (IDH2) PAK078Mu01 Fas Associating Death Domain Containing Protein (FADD) PAA537Hu01 Enolase, Neuron Specific (NSE) PAA651Hu01 Hyaluronan Binding Protein 1 (HABP1) PAB215Hu02 Fibrinogen Beta (FGb) PAB769Hu01 S100 Calcium Binding Protein A6 (S100A6) PAB231Hu01 Keratin 18 (KRT18) PAH839Hu01 Isocitrate Dehydrogenase 1, Soluble (IDH1) PAE748Hu01 Karyopherin Alpha 2 (KPNa2) PAB081Hu02 Heat Shock 70kDa Protein 1A (HSPA1A) PAA778Mu01 B-Cell Leukemia/Lymphoma 2 (Bcl2) PAA853Hu03 Caspase 8 (CASP8) PAA399Mu01 High Mobility Group Protein 1 (HMG1) PAA303Mu01 Galectin 3 (GAL3) PAA537Mu02 Enolase, Neuron Specific (NSE) PAA994Ra01 Acid Phosphatase 1 (ACP1) PAB083Ra01 Superoxide Dismutase 2, Mitochondrial (SOD2) PAB449Mu01 Enolase, Non Neuronal (NNE) PAA376Mu01 Actinin Alpha 2 (ACTN2) PAA553Ra01 Matrix Metalloproteinase 9 (MMP9) PAA929Bo01 Retinol Binding Protein 4, Plasma (RBP4) PAA491Ra02 Keratin 2 (KRT2) PAC025Hu01 Keratin 8 (KRT8) PAB231Mu01 Keratin 18 (KRT18) PAC598Hu03 Vanin 1 (VNN1) -

The Role of Signal Transducer and Activator of Transcription 5 in the Inhibitory Effects of GH on Adipocyte Differentiation
139 The role of signal transducer and activator of transcription 5 in the inhibitory effects of GH on adipocyte differentiation H E Richter1, T Albrektsen2 and N Billestrup1,3 1Department of Signal Transduction, Novo Nordisk A/S, Novo Allé, DK-2880 Bagsvaerd, Denmark 2Department of Transcription Biology, Novo Nordisk A/S, Novo Allé, DK-2880 Bagsvaerd, Denmark 3Steno Diabetes Centre, Niels Steensensvej 6, DK-2820 Gentofte, Denmark (Requests for offprints should be addressed to N Billestrup, Steno Diabetes Centre, Niels Steensensvej 6, NSK2·023, DK-2820 Gentofte, Denmark; Email:[email protected]) Abstract GH inhibits primary rat preadipocyte differentiation and expression of late genes required for terminal differentiation. Here we show that GH-mediated inhibition of fatty acid-binding protein aP2 gene expression correlates with the activation of the Janus kinase-2/signal transducer and activator of transcription (STAT)-5 signalling pathway. Within minutes of treatment, GH induced the tyrosine phosphorylation, nuclear localization and DNA binding of STAT5. Importantly, there was no evidence that STAT5 acted via an interaction with peroxisome proliferator-activated receptor γ. To further understand the mechanism of STAT5 action, we reconstituted the inhibition of aP2 in a non-adipogenic cell line. Using this system, we showed that the ability of GH to inhibit a 520 bp aP2 reporter was largely dependent upon the presence of either STAT5A or STAT5B. Mutant analysis confirmed that the tyrosine phosphorylation of STAT5 was essential for this signalling. However, STAT5’s C-terminal transactivation domain was fully dispensable for this inhibition. Taken together, these data confirm a key regulatory role of STAT5 in adipose tissue and point to STAT5 as the repressing modulator of GH-mediated inhibition in primary preadipocytes. -

Transcriptional and Post-Transcriptional Regulation of ATP-Binding Cassette Transporter Expression
Transcriptional and Post-transcriptional Regulation of ATP-binding Cassette Transporter Expression by Aparna Chhibber DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Pharmaceutical Sciences and Pbarmacogenomies in the Copyright 2014 by Aparna Chhibber ii Acknowledgements First and foremost, I would like to thank my advisor, Dr. Deanna Kroetz. More than just a research advisor, Deanna has clearly made it a priority to guide her students to become better scientists, and I am grateful for the countless hours she has spent editing papers, developing presentations, discussing research, and so much more. I would not have made it this far without her support and guidance. My thesis committee has provided valuable advice through the years. Dr. Nadav Ahituv in particular has been a source of support from my first year in the graduate program as my academic advisor, qualifying exam committee chair, and finally thesis committee member. Dr. Kathy Giacomini graciously stepped in as a member of my thesis committee in my 3rd year, and Dr. Steven Brenner provided valuable input as thesis committee member in my 2nd year. My labmates over the past five years have been incredible colleagues and friends. Dr. Svetlana Markova first welcomed me into the lab and taught me numerous laboratory techniques, and has always been willing to act as a sounding board. Michael Martin has been my partner-in-crime in the lab from the beginning, and has made my days in lab fly by. Dr. Yingmei Lui has made the lab run smoothly, and has always been willing to jump in to help me at a moment’s notice. -

Nuclear Hormone Receptor Antagonism with AP-1 by Inhibition of the JNK Pathway
Downloaded from genesdev.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway Carme Caelles,1 Jose´M. Gonza´lez-Sancho, and Alberto Mun˜oz2 Instituto de Investigaciones Biome´dicas, Consejo Superior de Investigaciones Cientı´ficas, E-28029 Madrid, Spain The activity of c-Jun, the major component of the transcription factor AP-1, is potentiated by amino-terminal phosphorylation on serines 63 and 73 (Ser-63/73). This phosphorylation is mediated by the Jun amino-terminal kinase (JNK) and required to recruit the transcriptional coactivator CREB-binding protein (CBP). AP-1 function is antagonized by activated members of the steroid/thyroid hormone receptor superfamily. Recently, a competition for CBP has been proposed as a mechanism for this antagonism. Here we present evidence that hormone-activated nuclear receptors prevent c-Jun phosphorylation on Ser-63/73 and, consequently, AP-1 activation, by blocking the induction of the JNK signaling cascade. Consistently, nuclear receptors also antagonize other JNK-activated transcription factors such as Elk-1 and ATF-2. Interference with the JNK signaling pathway represents a novel mechanism by which nuclear hormone receptors antagonize AP-1. This mechanism is based on the blockade of the AP-1 activation step, which is a requisite to interact with CBP. In addition to acting directly on gene transcription, regulation of the JNK cascade activity constitutes an alternative mode whereby steroids and retinoids may control cell fate and conduct their pharmacological actions as immunosupressive, anti-inflammatory, and antineoplastic agents. -

The Histone Acetylase PCAF Is a Nuclear Receptor Coactivator
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press The histone acetylase PCAF is a nuclear receptor coactivator Jorge C.G. Blanco,1,4 Saverio Minucci,1 Jianming Lu,1 Xiang-Jiao Yang,1 Kristen K. Walker,3 Hongwu Chen,3 Ronald M. Evans,2,3 Yoshihiro Nakatani,1 and Keiko Ozato1,5 1Laboratory of Molecular Growth Regulation, National Institute of Child Health and Human Development, National Institutes of Health (NIH), Bethesda, Maryland 20892-2753 USA; 2Howard Hughes Medical Institute; 3The Salk Institute for Biological Studies, La Jolla, California 92037 USA Whereas the histone acetylase PCAF has been suggested to be part of a coactivator complex mediating transcriptional activation by the nuclear hormone receptors, the physical and functional interactions between nuclear receptors and PCAF have remained unclear. Our efforts to clarify these relationships have revealed two novel properties of nuclear receptors. First, we demonstrate that the RXR/RAR heterodimer directly recruits PCAF from mammalian cell extracts in a ligand-dependent manner and that increased expression of PCAF leads to enhanced retinoid-responsive transcription. Second, we demonstrate that, in vitro, PCAF directly associates with the DNA-binding domain of nuclear receptors, independently of p300/CBP binding, therefore defining a novel cofactor interaction surface. Furthermore, our results show that dissociation of corepressors enables ligand-dependent PCAF binding to the receptors. This observation illuminates how a ligand-dependent receptor function can be propagated to regions outside the ligand-binding domain itself. On the basis of these observations, we suggest that PCAF may play a more central role in nuclear receptor function than previously anticipated. -

Mutant Thyroid Hormone Receptor Represses the Expression And
[CANCER RESEARCH 63, 5274–5280, September 1, 2003] Mutant Thyroid Hormone Receptor  Represses the Expression and Transcriptional Activity of Peroxisome Proliferator-activated Receptor ␥ during Thyroid Carcinogenesis Hao Ying, Hideyo Suzuki, Li Zhao, Mark C. Willingham, Paul Meltzer, and Sheue-Yann Cheng1 Laboratory of Molecular Biology, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland 20892-4264 [H. Y., H. S., L. Z., S-Y. C.]; National Human Genome Research Institute, NIH, Bethesda, Maryland 20892-4264 [P. M.]; and Department of Pathology, Wake Forest University School of Medicine, Winston-Salem, North Carolina 27157-1072 [M. C. W.] ABSTRACT not in 10 papillary carcinomas. This unique genetic rearrangement in follicular carcinoma was further confirmed by subsequent analyses The molecular genetics underlying thyroid carcinogenesis is not clear. using a larger number of samples (8). When fused to PAX8, PPAR␥1 Recent identification of a PAX8-peroxisome proliferator-activated receptor ␥ ␥ not only loses its capability to stimulate thiazolidinedione-induced (PPAR ) fusion gene in human thyroid follicular carcinoma suggests a ␥ tumor suppressor role of PPAR␥ in thyroid carcinogenesis. Mice harbor- transcription but also acts to inhibit PPAR 1 transcriptional activity ␥ ing a knockin mutant thyroid hormone  receptor (TRPV) spontaneously (7). However, how the loss of PPAR 1 transcriptional activity im- develop thyroid follicular carcinoma through pathological progression of pacts the normal functions of thyroid follicular cells is unclear. hyperplasia, capsular invasion, vascular invasion, anaplasia, and eventu- We have recently created a mutant mouse by targeting a mutation ally, distant organ metastasis. This mutant mouse (TRPV/PV mouse) (PV)totheTR gene locus (TRPV mice; Ref.