Situational Awareness and Forecasting for Norway
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tildeling Av Skjønnsmidler 2021 Trøndelag (Forhåndsfordelt Og 1
Vår dato: Vår ref: 19.11.2020 2020/7330 Deres dato: Deres ref: «REFDATO» «REF» «MOTTAKERNAVN» Saksbehandler, innvalgstelefon «ADRESSE» Sigrid Hynne, 74 16 80 79 «POSTNR» «POSTSTED» «KONTAKT» Tildeling av skjønnsmidler 2021 Trøndelag (forhåndsfordelt og 1. tildeling av tilbakeholdte midler) Trøndelag ble tildelt 98 mill. kr av den fylkesvise skjønnsrammen for 2021. På statsbudsjettkonferansen for Trøndelag som ble avholdt 8. oktober presenterte Fylkesmannen fordelingen av skjønnsmidler for 2020 og 2021. Dette brevet beskriver hva som så langt er fordelt til kommunene for 2021. Vedlegg 1 viser fordeling per kommune, mens vedlegg 2 inneholder oversikt over hvilke prosjekter som er tildelt skjønnsmidler for 2021 (fornying og innovasjon). Deler av skjønnsrammen vil holdes tilbake til fordeling gjennom 2021. I oktober neste år vil vi derfor komme tilbake med en samlet oppsummering av skjønnstildelingen for 2021. Skjønnsmidler 2020 For 2020-midlene viser vi til brev til KMD datert 15.10.2020, og som ble sendt i kopi til kommunene i fylket. På denne nettsiden ligger det en excel-fil som viser oversikt over fordeling av skjønn for 2020, der det er mulig å filtrere per kommune: https://www.fylkesmannen.no/nb/Trondelag/Kommunal- styring/Kommuneokonomi/Skjonnsmidler/ I tillegg vil vi i starten av desember sende et brev til kommunene om fordeling av ekstraordinære skjønnsmidler 2020 begrunnet i koronasituasjonen. Disse skjønnsmidlene blir utbetalt i desember. Om skjønnstildeling og retningslinjer Med bakgrunn i Kommunal- og moderniseringsdepartementets retningslinjer, og gjennom konsultasjon med kommunene, har Fylkesmannen i Trøndelag utviklet en tildelingsmodell med tilhørende retningslinjer (sist oppdatert mai 2020). Skjønnstildelingen bygger på hovedkomponentene kompensasjonsskjønn og prosjektskjønn, der prosjektskjønn dekker innovasjon og fornyingsprosjekter i enkeltkommuner og i samarbeid med flere, i tillegg til fellesløft (fellestiltak) for hele fylket. -

Planstrategi for Alver Kommune 2020-2023
Planstrategi for Alver kommune 2020-2023 1 1 Innleiing Kommunal planstrategi er eit verktøy for politisk prioritering av planinnsatsen, for å sikre ei ressurseffektiv og målretta planlegging. Planstrategien skal vedtakast av kommunestyret seinast eit år etter at det nye kommunestyret er konstituert. I tråd med Kommunal- og moderniseringsdepartementet si (KMD) tilråding har dei tre kommunane som blir Alver kommune frå 1.1.2020 vedteke å samordna planbehovet med heimel i PBL. § 9-1, før ny kommune formelt trer i kraft. Regjeringa legg kvart fjerde år fram nasjonale forventningar til regional og kommunal planlegging. Dei nasjonale forventningane skal fylgjast opp i fylkeskommunen og kommunane sitt arbeid med planstrategi og planer, og vert lagt til grunn for statlege myndigheter si medverknad i planlegginga. Regjeringa har bestemt at FN sine 17 berekraftmål, som Norge har slutta seg til, skal vere det politiske hovudsporet for å ta tak i vår tid sine største utfordringar. Det er difor viktig at berekraftmåla blir ein del av grunnlaget for samfunns- og arealplanlegginga. 2 Alver kommune 2.1 Mål og visjonar for Alver Alver kommune skal vera ein heilskapleg samfunnsutviklar og byggja fellesskap i ein region med sterk vekst og utvikling ( Intensjonsavtalen). 2.1.1 Verdiar, mål og prinsipp Følgjande er lagt til grunn i intensjonsavtalen for Alver kommune. • Me skal byggja ein ny kommune. • Alle kommunar er likeverdige partar. • Me skal ha respekt for kvarandre sine tradisjonar, kulturar og sterke eigenskapar i noverande organisasjon. • Me skal ta med oss beste praksis frå kommunane inn i den nye kommunen. • Me skal byggja vidare på felles identitet og kultur. -

Her Er Midt-Norges Beste Drikkevann - NRK Trøndelag - Lokale Nyheter, TV Og Radio Page 1 of 3
Her er Midt-Norges beste drikkevann - NRK Trøndelag - Lokale nyheter, TV og radio Page 1 of 3 TrøndelaJournalistg Her er Midt-Norges beste drikkevann Smak, lukt og farge var kriteriene da Midt-Norges beste drikkevann ble kåret. Innbyggerne på Frøya og i Oppdal kan nå konstatere at de har landsdelens beste drikkevann. Vinnerne kunne stolt motta premien for beste drikkevann. Fra venstre Ivar Meland og overflatevannet på Frøya. Til høyre Tore Samskott og grunnvannet i Oppdal. FOTO: BENT LINDSETMO / NRK Sigrun Hofstad Tom Erik Sørensen Publisert i dag, for 4 timer siden Onsdag formiddag ble Midt-Norges beste drikkevann kåret under Vann- og avløpsdagene på Stjørdal. Åtte regionvinnerne fra hele landet går til finalen i Telenor Arena 19. april 2016. Vinnerne i de to forskjellige klassene ble Frøya kommunale vannverk og Oppdal sentrum vannverk. Førstnevnte vant klassen for beste overflatevann, mens Oppdal kan smykke seg med tittelen i klassen for beste grunnvann eller borehullvann. – Det er ikke enkelt å kåre det beste vannet. Det er basert på smak, lukt og farge. Det er jo lett å se om vannet har farge, men smak og lukt blir jo en subjektiv vurdering, sier tidligere direktør i Norsk Vann, Einar Melheim. http://www.nrk.no/trondelag/her-er-midt-norges-beste-drikkevann-1.12625476 31.10.2015 Her er Midt-Norges beste drikkevann - NRK Trøndelag - Lokale nyheter, TV og radio Page 2 of 3 Det er Norsk Vann, Norsk Kommunalteknisk Forening og KS Bedrift som står bak konkurransen Norges beste drikkevann 2016. Formålet er å sette fokus på at drikkevann fra norske vannverk har høy kvalitet og koster en tusendel mindre enn flaskevann. -

Industrial and Commercial Parks in Greater Bergen
INDUSTRIAL AND COMMERCIAL PARKS IN GREATER BERGEN 1 Greater Bergen Home of the ocean industries Bergen and the surrounding munic- companies have shifted towards new ipalities are strategically located on green and sustainable solutions. Tone Hartvedt the west coast of Norway. Being that Vestland County, being the biggest Invest in Bergen close to the North Sea and its natural producer of hydropower in Norway, +47 917 29 055 resources has put the city and the makes the region an attractive place for [email protected] region in a unique position to take a establishing companies with business leading role in developing the ocean models based on hydropower. industries. The region is playing a significant role Vidar Totland Vestland County is the biggest county in developing new electric ferries and Invest in Bergen in Norway for exporting goods. is now leading in the construction of +47 959 12 970 Greater Bergen is home to the ocean boats fuelled by hydrogen. The new [email protected] industries, which are mainly based carbon capture and storage centre, on export. Shipping, new clean Northern Lights, is located here and maritime technology solutions, oil, will attract several other businesses gas, renewable energy, fishery, and to be part of the supply chain for this aquaculture are the leading ocean huge development. A green hydrogen industries in the region. production plant is also being planned for in the region. The green shift The region offers several possibilities Greater Bergen has the competence, for industries that want to transform sites, and spirit to be part of the green their businesses in a sustainable revolution we see coming. -

Endelig Tilsynsrapport Og Pålegg Om Utbedring
Returadresse: Arkivverket Pb.4013 Ullevål Stadion 0806 OSLO Indre Fosen kommune v/Rådmann Dato 26.03.2019 Rådhusveien 13 Din ref. 2018/11530 7100 RISSA Vår ref. 2018/20123 Saksbehandler THOOVE/DOFO Endelig tilsynsrapport og pålegg om utbedring Vi takker for godt samarbeid i forbindelse med tilsynet. Tilsynet hadde hovedfokus på arkivorganisering og elektronisk arkivdanning, blant annet arkivplan, journalføring og fagsystemer. Den endelige rapporten er justert i henhold til Indre Fosen kommune sin tilbakemelding av 12.3.2019. Tilsynsdato: 29.1.2019 Fra Indre Fosen Rådmann Vigdis Bolås kommune: Personal- og kommunikasjonssjef Ola Andreas Stavne Leder Fosen IKT Robert Karlsen Fagleder dokumentsenter Øyvind Lindseth Arkivkonsulent Rita Rosvoldaune Arkivmedarbeider Marte Bliksås Administrativ leder Fosen barneverntjeneste Mari Kilen Byggesaksbehandler Stian Fallrø Konsulent kultur/næring Ingunn Rokseth Rådmann, personal- og kommunikasjonssjef og leder for Fosen IKT var til stede under oppstartsmøtet og oppsummeringen. Byggesaksbehandler Fallrø og konsulent Rokseth deltok på intervjuer om arkivdanningen innenfor sine respektive fagområder. Administrativ leder for Fosen barneverntjeneste redegjorde for barneverntjenestens arkivhold. Med unntak for intervjuene med Fallrø og Rokseth var fagleder for dokumentsenter og arkivkonsulent Rosvoldaune til stede under hele tilsynet. Arkivmedarbeider Bliksås deltok på Organisasjonsnr.: 961181399 Telefon: 480 55 666 www.arkivverket.no/tjenester gjennomgangen av arkivplan og dokumentsenterets arbeid. Fra Arkivverket: Arkivar Thomas Øverby Seniorrådgiver Geir Ivar Tungesvik Positive funn fra tilsynsbesøket Tilsynet viser at kommunen har arbeidet godt med avsluttingen av sak-/arkivsystemene i Rissa og Leksvik og etableringen av nytt sak-/arkivsystem i Indre Fosen kommune. Saksbehandlerne som ble intervjuet under tilsynet uttalte at denne prosessen ikke har skapt problemer for deres arbeid og at de har enkel tilgang på dokumenter fra Rissa og Leksvik kommune som de har behov for i det daglige. -

TRONDHEIMSREGIONEN – En Arena for Samarbeid
TRONDHEIMSREGIONEN – en arena for samarbeid Bård Eidet, daglig leder Trondheimsregionen Hvorfor regionalt samarbeid? • For å løse oppgaver bedre og mer effektivt enn den enkelte kommune kan gjøre? • For å gi større kraft til felles interesser? • Fordi det er oppgaver som finner sine beste løsninger mellom en kommune og et fylkesnivå? Grunnleggende: Det må finnes en vilje til å samarbeide, og en felles forståelse av at fellesskapet tjener på det samlet sett …men hvorfor Trondheimsregionen? • Ole Eirik Almlid (adm.dir. i NHO) ved lanseringen av kommunebarometeret: • “Almlid mener målingen viser hvilke kommuner som kan oppleves som de mest attraktive for næringslivet. Han mener målingen viser at kommuner som ligger i randsonen til de store kommunene, og som definerer seg inn i storbyregionen framfor å vende blikket mot distriktskommuner i stedet, har en tendens til å score bra.” • Hva gjør Trondheimsregionen? Et samarbeid mellom 10 – nei 9 – nei 8 kommuner – politisk styrt Indre 4 programområder: Fosen Strategisk næringsutvikling Nye Interkommunal arealplan Profilering/kommunikasjon/attraktiv region Ledelse/samarbeid/interessepolitikk Ikke fokus på tjenesteproduksjon Det politiske organet . Åtte kommuner, fylkeskommunen som observatør Stjørdal, Malvik, Trondheim, Melhus, Midtre Gauldal, Orkdal, Skaun, Indre Fosen Styrke Trondheimsregionens utvikling i en nasjonal og internasjonal • konkurransesituasjon. Politiske hovedmål Strategisk næringsplan (SNP): * Øke BNP slik at den tilsvarer vår andel av befolkningen i 2020 * Doble antall teknologibedrifter og -arbeidsplasser innen 2025 Interkommunal arealplan (IKAP): * Klimavennlig arealbruk og transport * Boligbygging nær sentra og kollektivtilbud * Jordvern * Fordele veksten . Politisk idé * Vi oppnår mer sammen enn hver for oss Organisasjon Trondheimsregionen – regionrådet RR Trondheim kommune (ordfører, opposisjon, rådmann) vertskommune Arbeidsutvalget – AU Daglig leder (leder, nestleder + 2 ordførere) Prosjektleder Næringsrådet - NF med sekretariat for (2 FoU, 3 næringsliv, 3 næringsplanen. -

Taxi Midt-Norge, Trøndertaxi Og Vy Buss AS Skal Kjøre Fleksibel Transport I Regionene I Trøndelag Fra August 2021
Trondheim, 08.02.2021 Taxi Midt-Norge, TrønderTaxi og Vy Buss AS skal kjøre fleksibel transport i regionene i Trøndelag fra august 2021 Den 5. februar 2021 vedtok styret i AtB at Taxi Midt-Norge, TrønderTaxi og Vy Buss AS får tildelt kontraktene for fleksibel transport i Trøndelag fra august 2021. Transporttilbudet vil være med å utfylle rutetilbudet med buss. I tillegg er det tilpasset både regionbyer og distrikt, med servicetransport i lokalmiljøet og tilbringertransport for å knytte folk til det rutegående kollektivnettet med buss eller tog. Fleksibel transport betyr at kundene selv forhåndsbestiller en tur fra A til B basert på sitt reisebehov. Det er ikke knyttet opp mot faste rutetider eller faste ruter, men innenfor bestemte soner og åpningstider. Bestillingen skjer via bestillingsløsning i app, men kan også bestilles pr telefon. Fleksibel transport blir en viktig del av det totale kollektivtilbudet fra høsten 2021. Tilbudet er delt i 11 kontrakter. • Taxi Midt-Norge har vunnet 4 kontrakter og skal tilby fleksibel transport i Leka, Nærøysund, Grong, Høylandet, Lierne, Namsskogan, Røyrvik, Snåsa, Frosta, Inderøy og Levanger, deler av Steinkjer og Verdal, Indre Fosen, Osen, Ørland og Åfjord. • TrønderTaxi har vunnet 4 kontakter og skal tilby fleksibel transport i Meråker, Selbu, Tydal, Stjørdal, Frøya, Heim, Hitra, Orkland, Rindal, Melhus, Skaun, Midtre Gauldal, Oppdal og Rennebu. • Vy Buss skal drifte fleksibel transport tilpasset by på Steinkjer og Verdal, som er en ny og brukertilpasset måte å tilby transport til innbyggerne på, og som kommer i tillegg til rutegående tilbud med buss.Vy Buss vant også kontraktene i Holtålen, Namsos og Flatanger i tillegg til to pilotprosjekter for fleksibel transport i Røros og Overhalla, der målet er å utvikle framtidens mobilitetstilbud i distriktene, og service og tilbringertransport i områdene rundt disse pilotområdene. -

Mg:Nytt No 01 Juli 2016 Informasjonsavis for Midtre Gauldal Kommune
MG:NYTT NO 01 JULI 2016 INFORMASJONSAVIS FOR MIDTRE GAULDAL KOMMUNE KOMMUNESAMMENSLÅING ÅRETS KLIMASKOLE 2016 BRUKERUNDERSØKELSER God sommer! 2 | NO 01 JULI 2016 Ordfører Sivert Moen ha minst 42 prosent mere areal til bolig. Skal vi ha samme andel sysselsatte i egen kommune må det også etableres og utvikles næringsvirksomhet tilsvarende. Da må vi ha areal. Vi må ha nytt Sommeren er her! areal og det finner vi ikke i tilstrekkelig grad i Støren sentrum. Vi Sola har snudd og vi går mot den varmeste må opp av dalbotnen for å kunne vokse. Vi må opp av dalbotnen delen av sommeren. Vi har vært gjennom for å kunne ta vare på de verdier vi har her. Slik er det bare. en hektisk vinter og vår med mange tunge Hva er det som driver utvikling inn til kommunen vår på denne saker i kommunestyret. Kommunereformen måten? Det er i høy grad mangel på areal andre steder og at har vært en av de sakene hvor meningene Trondheimsregionen vokser. Ikke bare i befolkning, men også i har vært mange. Kommunestyret gikk til verdiskaping. Gjennom bedre kommunikasjoner blir avstandene slutt inn for å forhandle om en intensjonsav målt i tid kortet ned. Det planlegges for framtida med dobbeltspor tale med Melhus kommune om sammenslåing. Og der stoppet på jernbanen til Støren. Flytoget til Værnes vil bruke vel tre kvar det fordi Melhus ville forhandle med Skaun, og ikke med oss. Vi ter fra Støren. Likeledes blir avstanden fra Støren til Trondheim har brukt store ressurser og mye tid. Ikke har det vært så bortkas med ny firefelts motorveg på under 30minutter. -

NGT 60 3 175-188.Pdf
Geochronological investigations in the Oppdal area, central Norway STEINAR SOLHEIM Solheim, S.: Geochronological investigations in the Oppdal area, central Norway. Norsk Geologisk Tidsskrift, Vol. 60, pp. 175-188. Oslo 1980. ISSN 0029-196X. The isochron ages obtained have been grouped in four geological time periods: 1700-1750 m.y. (dioritic gneisses, GråhØ/TronfjeU Complexes), 1450-1500 m.y. (augen gneisses and intrusives, GråhØ/TronfjeU Complexes), -1050 m.y. (Basement Complex and Flagstone Group), and -400 m.y. (minerals). The Caledonian influence in the area seems to be smaller than previously assumed since it has on! y caused a resetting of the isotopic system between minerals. The isochron ages of- 1050 m.y. are thought to reflect secondary events during the Sveconorwegian Cycle. The time of deposition/intrusion of the rocks of the Basement Complex and the Flagstone Group is thought to be earlier than 1450-1500 m.y. One extensive metamorphism has been recognized, and 1450-1500 m.y. is considered to be a minimum age for this event. S. Solheim, Mineralogisk-geologisk museum, Sars gt. l, Oslo 5. Present address: Norsk Hydro, Oljedivisjonen, Kjørboko/len, 1300 Sandvika, Norway. The Oppdal area is situated in southern ses, granitic gneisses and augen gneisses. Eggen TrØndelag (Fig. l). Geologically the area is (1977) interprets the banded gneisses as bordered by Caledonian rocks of the Trondheim metavolcanics. In the southern part of the area Region in the east and Precambrian rocks of the the zone of outcrops of the banded gneisses Gneiss Region in the west. The aim of this work becomes narrower and it ends just south of the has been to date the various rock units in this area mapped by Eggen (1977), Evensen (1977) area and in this way to contribute to a hetter and Sjulsen (1977). -

Og Rådsmøte Fosen Regionråd 08.11.2019
FOSEN REGIONRÅD 7170 Åfjord - Tlf: 72 53 15 10 FRA: Styre- og rådet i Fosen Regionråd Sekretariatet/ TB Møtedato: 08.11.2019 STYRE- og RÅDSMØTE I FOSEN REGIONRÅD MØTEREFERAT Sted: «Møterom 4.etg Parken», Åfjord kommune Tid: Fredag 08.11.19, Kl 09:30 Deltakere: Det ble til dette møtet innkalt iht konstitueringene i kommunestyrene etter kommunevalget 2019, se liste under Gjester: Hallgeir Grøntvedt, leder hovedutvalg veg Trøndelag fylkeskommune Knut Sundet, daglig leder Fosenvegene AS Line Samuelsen, Trøndelag reiseliv Elisabeth Heyerdahl Refsum, Heyerdahl Refsum AS Rådet: Bjørnar Buhaug Indre Fosen kommune Steinar Saghaug Indre Fosen kommune Ivar Rostad Indre Fosen kommune John Einar Høvik Osen kommune Egil A. Johannessen Osen kommune Forfall, ingen vara i møtet Line Stein Osen kommune Forfall, ingen vara i møtet Tom Myrvold Ørland kommune Ogne Undertun Ørland kommune Finn Olav Odde Ørland kommune Forfall, ingen vara i møtet Vibeke Stjern Åfjord kommune Einar Eian Åfjord kommune Tone Bårdli Åfjord kommune Forfall, vara Erling Iversen deltok Rådmannsforum: Vigdis Bolås Indre Fosen kommune Roar Leirset Osen kommune Emil Raaen Ørland kommune Per Ola Johansen Åfjord kommune Øvrige deltakere; Ann-Berit Nervik Tillitsvalgt Leif Harald Hanssen Adm. representant Trøndelag fylkeskommune Stig Klomsten Pol. representant Forfall, ingen vara Trøndelag fylkeskommune Rune Schei Pol. representant Forfall, ingen vara Trøndelag fylkeskommune Frode Rabben Fylkesmannen i Forfall, ingen vara Trøndelag Torun Bakken Fosen Regionråd Oppmøte i styre- og rådsmøtet ifølge ovennevnte liste Møteleder: Styrets leder Ogne Undertun frem til valg. Etter valg av ny leder, Vibeke Stjern, ble møtet ledet av utviklingsleder Torun Bakken. SAK 22/19 Utviklingsprisen for Fosen 2018 og 2019 Styret/rådets vedtak: 1. -
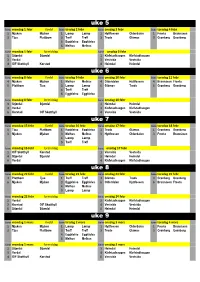
Uke 7 Uke 8 Uke 9 Uke 5 Uke 6
uke 5 bane mandag 1.febr Kveld bane tirsdag 2 febr bane onsdag 3 febr bane torsdag 4 febr 1 Mjuken Mjuken 1 Læmp Læmp 1 Hyttfossen Olderdalen 1 Frosta Brannåsen 3 Tjua Plattbom 3 Treff Treff 3 Trods Glåmos 3 Grønberg Grønberg 4 Eggkleiva Eggkleiva 6 Melhus Melhus bane mandag 1 febr formiddag bane onsdag 3 febr 1 Stjørdal Stjørdal 1 Klefstadhaugen Klefstadhaugen 3 Verdal 3 Vestsida Vestsida 4 OIF Skotthyll Korstad 4 Heimdal Heimdal uke 6 bane mandag 8 febr Kveld bane tirsdag 9 febr bane onsdag 10 febr bane torsdag 11 febr 4 Mjuken Mjuken 1 Melhus Melhus 4 Olderdalen Hyttfossen 4 Brannåsen Frosta 6 Plattbom Tjua 3 Læmp Læmp 6 Glåmos Trods 6 Grønberg Grønberg 4 Treff Treff 6 Eggkleiva Eggkleiva bane mandag 8 febr formiddag bane onsdag 10 febr 3 Stjørdal Stjørdal 1 Heimdal Heimdal 4 Verdal 3 Klefstadhaugen Klefstadhaugen 6 Korstad OIF Skotthyll 4 Vestsida Vestsida uke 7 bane mandag 15 febr Kveld bane tirsdag 16 febr bane onsdag 17 febr bane torsdag 18 febr 1 Tjua Plattbom 1 Eggkleiva Eggkleiva 1 Trods Glåmos 1 Grønberg Grønberg 3 Mjuken Mjuken 3 Melhus Melhus 3 Hyttfossen Olderdalen 3 Frosta Brannåsen 4 Læmp Læmp 6 Treff Treff bane mandag 15 febr formiddag bane onsdag 17 febr 1 OIF Skotthyll Korstad 1 Vestsida Vestsida 4 Stjørdal Stjørdal 3 Heimdal Heimdal 6 Verdal 4 Klefstadhaugen Klefstadhaugen uke 8 bane mandag 22 febr Kveld bane tirsdag 23 febr bane onsdag 24 febr bane torsdag 25 febr 4 Plattbom Tjua 1 Treff Treff 4 Glåmos Trods 4 Grønberg Grønberg 6 Mjuken Mjuken 3 Eggkleiva Eggkleiva 6 Olderdalen Hyttfossen 6 Brannåsen Frosta 4 Melhus -

University of London Deviant Burials in Viking-Age
UNIVERSITY OF LONDON DEVIANT BURIALS IN VIKING-AGE SCANDINAVIA Ruth Lydia Taylor M. Phil, Institute of Archaeology, University College London UMI Number: U602472 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. Dissertation Publishing UMI U602472 Published by ProQuest LLC 2014. Copyright in the Dissertation held by the Author. Microform Edition © ProQuest LLC. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 ABSTRACT DEVIANT BURIALS IN VIKING-AGE SCANDINAVIA The thesis brings together information yielded from archaeology and other sources to provide an overall picture of the types of burial practices encountered during the Viking-Age in Scandinavia. From this, an attempt is made to establish deviancy. Comparative evidence, such as literary, runic, legal and folkloric evidence will be used critically to shed perspective on burial practices and the artefacts found within the graves. The thesis will mostly cover burials from the Viking Age (late 8th century to the mid- 11th century), but where the comparative evidence dates from other periods, its validity is discussed accordingly. Two types of deviant burial emerged: the criminal and the victim. A third type, which shows distinctive irregularity yet lacks deviancy, is the healer/witch burial.