University of Southampton Research Repository Eprints Soton
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

User Office, Proposal Handling and Analysis Software
NOBUGS2002/031 October 11, 2002 Session Key: PB-1 User office, proposal handling and analysis software Jörn Beckmann, Jürgen Neuhaus Technische Universität München ZWE FRM-II D-85747 Garching, Germany [email protected] At FRM-II the installation of a user office software is under consideration, supporting tasks like proposal handling, beam time allocation, data handling and report creation. Although there are several software systems in use at major facilities, most of them are not portable to other institutes. In this paper the requirements for a modular and extendable user office software are discussed with focus on security related aspects like how to prevent a denial of service attack on fully automated systems. A suitable way seems to be the creation of a web based application using Apache as webserver, MySQL as database system and PHP as scripting language. PRESENTED AT NOBUGS 2002 Gaithersburg, MD, U.S.A, November 3-5, 2002 1 Requirements of user office software At FRM-II a user office will be set up to deal with all aspects of business with guest scientists. The main part is the handling and review of proposals, allocation of beam time, data storage and retrieval, collection of experimental reports and creation of statistical reports about facility usage. These requirements make off-site access to the user office software necessary, raising some security concerns which will be addressed in chapter 2. The proposal and beam time allocation process is planned in a way that scientists draw up a short description of the experiment including a review of the scientific background and the impact results from the planned experiment might have. -

An Interdisciplinary Journal
FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITA LISM FAST CAPITALISMFast Capitalism FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM ISSNFAST XXX-XXXX CAPITALISM FAST Volume 1 • Issue 1 • 2005 CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITA LISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITA LISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITA LISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITA LISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM FAST CAPITA LISM FAST CAPITALISM FAST CAPITALISM FAST CAPITALISM -
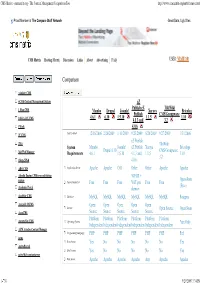
CMS Matrix - Cmsmatrix.Org - the Content Management Comparison Tool
CMS Matrix - cmsmatrix.org - The Content Management Comparison Tool http://www.cmsmatrix.org/matrix/cms-matrix Proud Member of The Compare Stuff Network Great Data, Ugly Sites CMS Matrix Hosting Matrix Discussion Links About Advertising FAQ USER: VISITOR Compare Search Return to Matrix Comparison <sitekit> CMS +CMS Content Management System eZ Publish eZ TikiWiki 1 Man CMS Mambo Drupal Joomla! Xaraya Bricolage Publish CMS/Groupware 4.6.1 6.10 1.5.10 1.1.5 1.10 1024 AJAX CMS 4.1.3 and 3.2 1Work 4.0.6 2F CMS Last Updated 12/16/2006 2/26/2009 1/11/2009 9/23/2009 8/20/2009 9/27/2009 1/31/2006 eZ Publish 2flex TikiWiki System Mambo Joomla! eZ Publish Xaraya Bricolage Drupal 6.10 CMS/Groupware 360 Web Manager Requirements 4.6.1 1.5.10 4.1.3 and 1.1.5 1.10 3.2 4Steps2Web 4.0.6 ABO.CMS Application Server Apache Apache CGI Other Other Apache Apache Absolut Engine CMS/news publishing 30EUR + system Open-Source Approximate Cost Free Free Free VAT per Free Free (Free) Academic Portal domain AccelSite CMS Database MySQL MySQL MySQL MySQL MySQL MySQL Postgres Accessify WCMS Open Open Open Open Open License Open Source Open Source AccuCMS Source Source Source Source Source Platform Platform Platform Platform Platform Platform Accura Site CMS Operating System *nix Only Independent Independent Independent Independent Independent Independent ACM Ariadne Content Manager Programming Language PHP PHP PHP PHP PHP PHP Perl acms Root Access Yes No No No No No Yes ActivePortail Shell Access Yes No No No No No Yes activeWeb contentserver Web Server Apache Apache -

Using Wordpress to Manage Geocontent and Promote Regional Food Products
Farm2.0: Using Wordpress to Manage Geocontent and Promote Regional Food Products Amenity Applewhite Farm2.0: Using Wordpress to Manage Geocontent and Promote Regional Food Products Dissertation supervised by Ricardo Quirós PhD Dept. Lenguajes y Sistemas Informaticos Universitat Jaume I, Castellón, Spain Co-supervised by Werner Kuhn, PhD Institute for Geoinformatics Westfälische Wilhelms-Universität, Münster, Germany Miguel Neto, PhD Instituto Superior de Estatística e Gestão da Informação Universidade Nova de Lisboa, Lisbon, Portugal March 2009 Farm2.0: Using Wordpress to Manage Geocontent and Promote Regional Food Products Abstract Recent innovations in geospatial technology have dramatically increased the utility and ubiquity of cartographic interfaces and spatially-referenced content on the web. Capitalizing on these developments, the Farm2.0 system demonstrates an approach to manage user-generated geocontent pertaining to European protected designation of origin (PDO) food products. Wordpress, a popular open-source publishing platform, supplies the framework for a geographic content management system, or GeoCMS, to promote PDO products in the Spanish province of Valencia. The Wordpress platform is modified through a suite of plug-ins and customizations to create an extensible application that could be easily deployed in other regions and administrated cooperatively by distributed regulatory councils. Content, either regional recipes or map locations for vendors and farms, is available for syndication as a GeoRSS feed and aggregated with outside feeds in a dynamic web map. To Dad, Thanks for being 2TUF: MTLI 4 EVA. Acknowledgements Without encouragement from Dr. Emilio Camahort, I never would have had the confidence to ensure my thesis handled the topics I was most passionate about studying - sustainable agriculture and web mapping. -

UC Irvine UC Irvine Previously Published Works
UC Irvine UC Irvine Previously Published Works Title Dimethyl Fumarate Alleviates Dextran Sulfate Sodium-Induced Colitis, through the Activation of Nrf2-Mediated Antioxidant and Anti-inflammatory Pathways. Permalink https://escholarship.org/uc/item/7n185057 Journal Antioxidants (Basel, Switzerland), 9(4) ISSN 2076-3921 Authors Li, Shiri Takasu, Chie Lau, Hien et al. Publication Date 2020-04-24 DOI 10.3390/antiox9040354 Peer reviewed eScholarship.org Powered by the California Digital Library University of California antioxidants Article Dimethyl Fumarate Alleviates Dextran Sulfate Sodium-Induced Colitis, through the Activation of Nrf2-Mediated Antioxidant and Anti-nflammatory Pathways Shiri Li 1, Chie Takasu 1 , Hien Lau 1, Lourdes Robles 1, Kelly Vo 1, Ted Farzaneh 2, Nosratola D. Vaziri 3, Michael J. Stamos 1 and Hirohito Ichii 1,* 1 Department of Surgery, University of California, Irvine, CA 92868, USA; [email protected] (S.L.); [email protected] (C.T.); [email protected] (H.L.); [email protected] (L.R.); [email protected] (K.V.); [email protected] (M.J.S.) 2 Department of Pathology, University of California, Irvine, CA 92868, USA; [email protected] 3 Department of Medicine, University of California, Irvine, CA 92868, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-714-456-8590; Fax: +1-714-456-8796 Received: 14 March 2020; Accepted: 22 April 2020; Published: 24 April 2020 Abstract: Oxidative stress and chronic inflammation play critical roles in the pathogenesis of ulcerative colitis (UC) and inflammatory bowel diseases (IBD). A previous study has demonstrated that dimethyl fumarate (DMF) protects mice from dextran sulfate sodium (DSS)-induced colitis via its potential antioxidant capacity, and by inhibiting the activation of the NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome. -

Open Source Katalog 2009 – Seite 1
Optaros Open Source Katalog 2009 – Seite 1 OPEN SOURCE KATALOG 2009 350 Produkte/Projekte für den Unternehmenseinsatz OPTAROS WHITE PAPER Applikationsentwicklung Assembly Portal BI Komponenten Frameworks Rules Engine SOA Web Services Programmiersprachen ECM Entwicklungs- und Testumgebungen Open Source VoIP CRM Frameworks eCommerce BI Infrastrukturlösungen Programmiersprachen ETL Integration Office-Anwendungen Geschäftsanwendungen ERP Sicherheit CMS Knowledge Management DMS ESB © Copyright 2008. Optaros Open Source Katalog 2009 - Seite 2 Optaros Referenz-Projekte als Beispiele für Open Source-Einsatz im Unternehmen Kunde Projektbeschreibung Technologien Intranet-Plattform zur Automatisierung der •JBossAS Geschäftsprozesse rund um „Information Systems •JBossSeam Compliance“ •jQuery Integrationsplattform und –architektur NesOA als • Mule Enterprise Bindeglied zwischen Vertriebs-/Service-Kanälen und Service Bus den Waren- und Logistiksystemen •JBossMiddleware stack •JBossMessaging CRM-Anwendung mit Fokus auf Sales-Force- •SugarCRM Automation Online-Community für die Entwickler rund um die •AlfrescoECM Endeca-Search-Software; breit angelegtes •Liferay Enterprise Portal mit Selbstbedienungs-, •Wordpress Kommunikations- und Diskussions-Funktionalitäten Swisscom Labs: Online-Plattform für die •AlfrescoWCMS Bereitstellung von zukünftigen Produkten (Beta), •Spring, JSF zwecks Markt- und Early-Adopter-Feedback •Nagios eGovernment-Plattform zur Speicherung und •AlfrescoECM Zurverfügungstellung von Verwaltungs- • Spring, Hibernate Dokumenten; integriert -

Sophie's World
Sophie’s World Jostien Gaarder Reviews: More praise for the international bestseller that has become “Europe’s oddball literary sensation of the decade” (New York Newsday) “A page-turner.” —Entertainment Weekly “First, think of a beginner’s guide to philosophy, written by a schoolteacher ... Next, imagine a fantasy novel— something like a modern-day version of Through the Looking Glass. Meld these disparate genres, and what do you get? Well, what you get is an improbable international bestseller ... a runaway hit... [a] tour deforce.” —Time “Compelling.” —Los Angeles Times “Its depth of learning, its intelligence and its totally original conception give it enormous magnetic appeal ... To be fully human, and to feel our continuity with 3,000 years of philosophical inquiry, we need to put ourselves in Sophie’s world.” —Boston Sunday Globe “Involving and often humorous.” —USA Today “In the adroit hands of Jostein Gaarder, the whole sweep of three millennia of Western philosophy is rendered as lively as a gossip column ... Literary sorcery of the first rank.” —Fort Worth Star-Telegram “A comprehensive history of Western philosophy as recounted to a 14-year-old Norwegian schoolgirl... The book will serve as a first-rate introduction to anyone who never took an introductory philosophy course, and as a pleasant refresher for those who have and have forgotten most of it... [Sophie’s mother] is a marvelous comic foil.” —Newsweek “Terrifically entertaining and imaginative ... I’ll read Sophie’s World again.” — Daily Mail “What is admirable in the novel is the utter unpretentious-ness of the philosophical lessons, the plain and workmanlike prose which manages to deliver Western philosophy in accounts that are crystal clear. -

Making Where Available to the Community
Making Where Available to the Community Geir Arne Hjelle, Ann-Silje Kirkvik, Michael Dahnn,¨ Ingrid Fausk Abstract The Norwegian Mapping Authority is an as- extend the capabilities of the software going forward, sociated Analysis Center within the IVS and is cur- including preliminary support for SLR and GNSS anal- rently preparing to contribute VLBI analyses to the yses. IVS with its new Where software. The software is be- The background for why NMA has developed a ing made available to the geodetic community under new software and which models are available, as well an open source MIT license. You can download Where as early benchmarks and results, are covered in [3] from GitHub and try it for yourself. Where is written and [4]. This note will focus more on how you can ob- in Python and comes with a graphical tool called There tain and use the software yourself. that can be used to analyze results from Where and edit sessions. Furthermore, a geodetic Python library called Midgard is made available. This library contains 2 Overview of Where components of Where that are useful in a wider setting. If you are doing any geodetic data analysis in Python, 1 Midgard might be useful to you. The Where software is mainly written in Python . It is cross platform and can run on Linux, macOS, and Windows. Where is available for download at Keywords VLBI, Software, Python, Open source kartverket.github.io/where/ Where can do different kinds of analyses. Each kind of analysis is defined as a pipeline consisting of sepa- 1 Introduction rate steps or stages. -
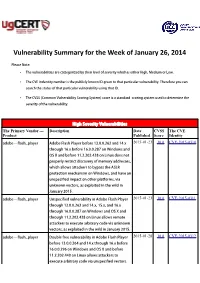
Vulnerability Summary for the Week of January 26, 2014
Vulnerability Summary for the Week of January 26, 2014 Please Note: • The vulnerabilities are cattegorized by their level of severity which is either High, Medium or Low. • The !" indentity number is the #ublicly $nown %& given to that #articular vulnerability. Therefore you can search the status of that #articular vulnerability using that %&. • The !'S (Common !ulnerability 'coring System) score is a standard scoring system used to determine the severity of the vulnerability. High Severity Vulnerabilities The Primary Vendor --- Description Date CVSS The CVE Product Published Score Identity adobe ** flash+#layer ,dobe -lash Player before ./.0.0.121 and .3.x 2015-01-23 10.0 CVE-2015-0310 through .2.x before .2.0.0.256 on 7indows and 8' 9 and before ...2.201.4/5 on Linu4 does not #ro#erly restrict discovery of memory addresses, which allows attac$ers to bypass the ,'L: #rotection mechanism on 7indows, and have an uns#ecified im#act on other #latforms, via un$nown vectors, as e4#loited in the wild in ;anuary 10.<. adobe ** flash+#layer =ns#ecified vulnerability in ,dobe -lash Player 2015-01-23 10.0 CVE-2015-0311 through ./.0.0.221 and .3.x, .<.x, and .2.x through .2.0.0.256 on 7indows and 8' 9 and through ...1.201.4/5 on Linu4 allows remote attac$ers to e4ecute arbitrary code via un$nown vectors, as e4#loited in the wild in ;anuary 10.<. adobe ** flash+#layer Double free vulnerability in ,dobe -lash Player 2015-01-28 10.0 CVE-2015-0312 before ./.0.0.123 and .3.x through .2.x before .2.0.0.2>2 on 7indows and 8' 9 and before ...2.101.430 on Linu4 allows attac$ers to e4ecute arbitrary code via uns#ecified vectors. -
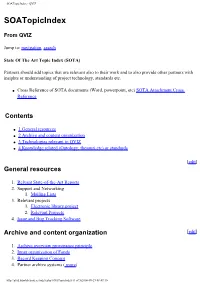
Soatopicindex - QVIZ
SOATopicIndex - QVIZ SOATopicIndex From QVIZ Jump to: navigation, search State Of The Art Topic Index (SOTA) Partners should add topics that are relevant also to their work and to also provide other partners with insights or understanding of project technology, standards etc. ● Cross Reference of SOTA documents (Word, powerpoint, etc) SOTA Attachment Cross Reference Contents ● 1 General resources ● 2 Archive and content organization ● 3 Technologies relevant to QVIZ ● 4 Knowledge related (Ontology, thesauri,etc) or standards [edit] General resources 1. Relvant State-of-the-Art Reports 2. Support and Networking 1. Mailing Lists 3. Relevant projects 1. Electronic library project 2. Relevant Projects 4. Issue and Bug Tracking Software Archive and content organization [edit] 1. Archive overview provenance principle 2. Inner organization of Fonds 3. Record Keeping Concept 4. Partner archive systems ( more) http://qviz.humlab.umu.se/index.php/SOATopicIndex (1 of 5)2006-09-29 08:49:10 SOATopicIndex - QVIZ 1. NAE System Description 2. SVAR and National Archives System Description 3. Vision of Britain System Description 4. Comparison of admin unit issues across partners systems 5. Archive features - QVIZ 1. Trackback 6. Archive Standards Technologies relevant to QVIZ [edit] 1. Image annotation to support user generated Thematic maps 2. Web Tools: Screen Capture 3. Prominent digital repositories Technologies and Digital Archive Technologies 1. See also Digital Object Metadata 4. Semantic repositories and other basis repository techologies (also semantic digital or semantic e-Libraries, etc) 5. Semantic web services (SWS) and Service oriented architecture (SOA) 6. Access Stategies 7. Workflow Technologies (includes BEPL,etc related tools) 8. Relevant social software (mainly to point out relevant features) 1. -

Arcane Wonders Ape Games Ares Games Alliance Game
GAMES ALLIANCE GAME DISTRIBUTORS ARES GAMES DIVINITY DERBY Ready to place bets on a race of mythological proportions? Zeus has invited you, along with other divine friends from across the Multiverse, for GAMES a little get–together on Mount Olympus. After a legendary lunch and a few pints of Ambrosia, you start wagering bets on which mythic creature is the fastest - and the only way to know for GAME TRADE MAGAZINE #211 certain is to summon all those creatures GTM contains articles on gameplay, for an epic race, with the Olympic “All- previews and reviews, game related father”, Zeus, as the ultimate judge! fiction, and self contained games and In Divinity Derby, players assume the game modules, along with solicitation role of a god (Anansi, Horus, Marduk, information on upcoming game and Odin, Quetzalcoatl, and Yu Huang the hobby supply releases. Jade Emperor), betting on a race among GTM 211 ...................................$3.99 NOT FINAL ART mythological flying creatures. Scheduled to ship in September 2017. AGS AREU004 .............................................................................................. $39.90 ART FROM PREVIOUS ISSUE GALAXY DEFENDERS: FINAL COUNTDOWN The end of the war is imminent - let’s start the Final Countdown! Expand the Galaxy Defenders agency’s army with APE GAMES these powerful, new agents - Scandium, Vanadium, and Xeno-Warrior - and unleash their unique classes, powers, and items! Plus, take the battlefield to the Third Dimension with new Doors and Windows Stand-up tokens! Also included is an additional set of custom Galaxy Defenders dice. Scheduled to ship in September 2017. AGS GRPR008 ............................. $39.90 LAST FRIDAY: RETURN TO CAMP APACHE Only Evil Can Stop Evil! In the DARK IS THE NIGHT wake of the tragic Camp Apache massacre over a decade ago, an Hunt or be Hunted! A hunter has set up ancient Evil is awakening - again - camp in a dark and dangerous forest. -

July/August 2021
July/August 2021 A Straight Path to the FreeBSD Desktop Human Interface Device (HID) Support in FreeBSD 13 The Panfrost Driver Updating FreeBSD from Git ® J O U R N A L LETTER E d i t o r i a l B o a r d from the Foundation John Baldwin FreeBSD Developer and Chair of ne of the myths surrounding FreeBSD is that it • FreeBSD Journal Editorial Board. is only useful in server environments or as the Justin Gibbs Founder of the FreeBSD Foundation, • President of the FreeBSD Foundation, foundation for appliances. The truth is FreeBSD and a Software Engineer at Facebook. O is also a desktop operating system. FreeBSD’s base sys- Daichi Goto Director at BSD Consulting Inc. tem and packages include device drivers for modern • (Tokyo). graphics adapters and input devices. Consistent with Tom Jones FreeBSD Developer, Internet Engineer FreeBSD’s role as a toolkit, FreeBSD supports a variety • and Researcher at the University of Aberdeen. of graphical interfaces ranging from minimalist window managers to full-featured desktop environments. The Dru Lavigne Author of BSD Hacks and • The Best of FreeBSD Basics. first article in this issue walks through several of these Michael W Lucas Author of more than 40 books including options explaining how users can tailor their desktop • Absolute FreeBSD, the FreeBSD to their needs. It also provides pointers to downstream Mastery series, and git commit murder. projects which build an integrated desktop system on Ed Maste Senior Director of Technology, top of FreeBSD. The next two articles dig into the details • FreeBSD Foundation and Member of the FreeBSD Core Team.