Molecular Basis of AMP Deaminase Deficiency in Skeletal Muscle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

35 Disorders of Purine and Pyrimidine Metabolism
35 Disorders of Purine and Pyrimidine Metabolism Georges van den Berghe, M.- Françoise Vincent, Sandrine Marie 35.1 Inborn Errors of Purine Metabolism – 435 35.1.1 Phosphoribosyl Pyrophosphate Synthetase Superactivity – 435 35.1.2 Adenylosuccinase Deficiency – 436 35.1.3 AICA-Ribosiduria – 437 35.1.4 Muscle AMP Deaminase Deficiency – 437 35.1.5 Adenosine Deaminase Deficiency – 438 35.1.6 Adenosine Deaminase Superactivity – 439 35.1.7 Purine Nucleoside Phosphorylase Deficiency – 440 35.1.8 Xanthine Oxidase Deficiency – 440 35.1.9 Hypoxanthine-Guanine Phosphoribosyltransferase Deficiency – 441 35.1.10 Adenine Phosphoribosyltransferase Deficiency – 442 35.1.11 Deoxyguanosine Kinase Deficiency – 442 35.2 Inborn Errors of Pyrimidine Metabolism – 445 35.2.1 UMP Synthase Deficiency (Hereditary Orotic Aciduria) – 445 35.2.2 Dihydropyrimidine Dehydrogenase Deficiency – 445 35.2.3 Dihydropyrimidinase Deficiency – 446 35.2.4 Ureidopropionase Deficiency – 446 35.2.5 Pyrimidine 5’-Nucleotidase Deficiency – 446 35.2.6 Cytosolic 5’-Nucleotidase Superactivity – 447 35.2.7 Thymidine Phosphorylase Deficiency – 447 35.2.8 Thymidine Kinase Deficiency – 447 References – 447 434 Chapter 35 · Disorders of Purine and Pyrimidine Metabolism Purine Metabolism Purine nucleotides are essential cellular constituents 4 The catabolic pathway starts from GMP, IMP and which intervene in energy transfer, metabolic regula- AMP, and produces uric acid, a poorly soluble tion, and synthesis of DNA and RNA. Purine metabo- compound, which tends to crystallize once its lism can be divided into three pathways: plasma concentration surpasses 6.5–7 mg/dl (0.38– 4 The biosynthetic pathway, often termed de novo, 0.47 mmol/l). starts with the formation of phosphoribosyl pyro- 4 The salvage pathway utilizes the purine bases, gua- phosphate (PRPP) and leads to the synthesis of nine, hypoxanthine and adenine, which are pro- inosine monophosphate (IMP). -

Metabolism of Purines and Pyrimidines in Health and Disease
39th Meeting of the Polish Biochemical Society Gdañsk 16–20 September 2003 SESSION 6 Metabolism of purines and pyrimidines in health and disease Organized by A. C. Sk³adanowski, A. Guranowski 182 Session 6. Metabolism of purines and pyrimidines in health and disease 2003 323 Lecture The role of DNA methylation in cytotoxicity mechanism of adenosine analogues in treatment of leukemia Krystyna Fabianowska-Majewska Zak³ad Chemii Medycznej IFiB, Uniwersytet Medyczny, ul. Mazowiecka 6/8, 92 215 £ódŸ Changes in DNA methylation have been recognized tory effects of cladribine and fludarabine on DNA as one of the most common molecular alterations in hu- methylation, after 48 hr growth of K562 cells with the man neoplastic diseases and hypermethylation of drugs, are non-random and affect mainly CpG rich is- gene-promoter regions is one of the most frequent lands or CCGG sequences but do not affect sepa- mechanisms of the loss of gene functions. For this rea- rately-located CpG sequences. The analysis showed son, DNA methylation may be a tool for detection of that cladribine (0.1 mM) reduced the methylated early cell transformations as well as predisposition to cytosines in CpG islands and CCGG sequences to a sim- metastasis process. Moreover, DNA methylation seems ilar degree. The inhibition of cytosine methylation by to be a promissing target for new preventive and thera- fludarabine (3 mM) was observed mainly in CCGG se- peutic strategies. quences, sensitive to HpaII, but the decline in the meth- Our studies on DNA methylation and cytotoxicity ylated cytosine, located in CpG island was 2-fold lower mechanism of antileukemic drugs, cladribine and than that with cladribine. -

Determining HDAC8 Substrate Specificity by Noah Ariel Wolfson A
Determining HDAC8 substrate specificity by Noah Ariel Wolfson A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biological Chemistry) in the University of Michigan 2014 Doctoral Committee: Professor Carol A. Fierke, Chair Professor Robert S. Fuller Professor Anna K. Mapp Associate Professor Patrick J. O’Brien Associate Professor Raymond C. Trievel Dedication My thesis is dedicated to all my family, mentors, and friends who made getting to this point possible. ii Table of Contents Dedication ....................................................................................................................................... ii List of Figures .............................................................................................................................. viii List of Tables .................................................................................................................................. x List of Appendices ......................................................................................................................... xi Abstract ......................................................................................................................................... xii Chapter 1 HDAC8 substrates: Histones and beyond ...................................................................... 1 Overview ..................................................................................................................................... 1 HDAC introduction -

TITLE Adenylate Kinase 2 Deficiency Causes NAD+ Depletion and Impaired Purine Metabolism During Myelopoiesis
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.05.450633; this version posted July 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. TITLE Adenylate Kinase 2 deficiency causes NAD+ depletion and impaired purine metabolism during myelopoiesis AUTHORS Wenqing Wang1, Andew DeVilbiss2, Martin Arreola1, Thomas Mathews2, Misty Martin-Sandoval2, Zhiyu Zhao2, Avni Awani1, Daniel Dever1, Waleed Al-Herz3, Luigi Noratangelo4, Matthew H. Porteus1, Sean J. Morrison2, Katja G. Weinacht1, * 1. Department of Pediatrics, Stanford University School of Medicine, Stanford, California 94305 USA 2. Children’s Research Institute, University of Texas Southwestern Medical Center, Dallas, Texas 75390 USA 3. Department of Pediatrics, Faculty of Medicine, Kuwait University, Safat, 13110 Kuwait 4. Laboratory of Clinical Immunology and Microbiology, National Institute of Health, BETHESDA MD 20814 USA * Corresponding author ABSTRACT Reticular Dysgenesis is a particularly grave from of severe combined immunodeficiency (SCID) that presents with severe congenital neutropenia and a maturation arrest of most cells of the lymphoid lineage. The disease is caused by biallelic loss of function mutations in the mitochondrial enzyme Adenylate Kinase 2 (AK2). AK2 mediates the phosphorylation of adenosine monophosphate (AMP) to adenosine diphosphate (ADP) as substrate for adenosine triphosphate (ATP) synthesis in the mitochondria. Accordingly, it has long been hypothesized that a decline in OXPHOS metabolism is the driver of the disease. The mechanistic basis for Reticular Dysgenesis, however, remained incompletely understood, largely due to lack of appropriate model systems to phenocopy the human disease. -

Role of the HPRG Component of Striated Muscle AMP Deaminase in the Stability and Cellular Behaviour of the Enzyme
biomolecules Review Role of the HPRG Component of Striated Muscle AMP Deaminase in the Stability and Cellular Behaviour of the Enzyme Francesca Ronca * and Antonio Raggi Laboratory of Biochemistry, Department of Pathology, University of Pisa, via Roma 55, 56126 Pisa, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-050-2218-273; Fax: +39-050-2218-660 Received: 19 July 2018; Accepted: 20 August 2018; Published: 23 August 2018 Abstract: Multiple muscle-specific isoforms of the Zn2+ metalloenzyme AMP deaminase (AMPD) have been identified based on their biochemical and genetic differences. Our previous observations suggested that the metal binding protein histidine-proline-rich glycoprotein (HPRG) participates in the assembly and maintenance of skeletal muscle AMP deaminase (AMPD1) by acting as a zinc chaperone. The evidence of a role of millimolar-strength phosphate in stabilizing the AMPD-HPRG complex of both AMPD1 and cardiac AMP deaminase (AMPD3) is suggestive of a physiological mutual dependence between the two subunit components with regard to the stability of the two isoforms of striated muscle AMPD. The observed influence of the HPRG content on the catalytic behavior of the two enzymes further strengthens this hypothesis. Based on the preferential localization of HPRG at the sarcomeric I-band and on the presence of a Zn2+ binding motif in the N-terminal regions of fast TnT and of the AMPD1 catalytic subunit, we advance the hypothesis that the Zn binding properties of HPRG could promote the association of AMPD1 to the thin filament. Keywords: AMP deaminase (AMPD); histidine-proline-rich glycoprotein (HPRG); striated muscle; Troponin T (TnT) 1. -

Developmental Disorder Associated with Increased Cellular Nucleotidase Activity (Purine-Pyrimidine Metabolism͞uridine͞brain Diseases)
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 11601–11606, October 1997 Medical Sciences Developmental disorder associated with increased cellular nucleotidase activity (purine-pyrimidine metabolismyuridineybrain diseases) THEODORE PAGE*†,ALICE YU‡,JOHN FONTANESI‡, AND WILLIAM L. NYHAN‡ Departments of *Neurosciences and ‡Pediatrics, University of California at San Diego, La Jolla, CA 92093 Communicated by J. Edwin Seegmiller, University of California at San Diego, La Jolla, CA, August 7, 1997 (received for review June 26, 1997) ABSTRACT Four unrelated patients are described with a represent defects of purine metabolism, although no specific syndrome that included developmental delay, seizures, ataxia, enzyme abnormality has been identified in these cases (6). In recurrent infections, severe language deficit, and an unusual none of these disorders has it been possible to delineate the behavioral phenotype characterized by hyperactivity, short mechanism through which the enzyme deficiency produces the attention span, and poor social interaction. These manifesta- neurological or behavioral abnormalities. Therapeutic strate- tions appeared within the first few years of life. Each patient gies designed to treat the behavioral and neurological abnor- displayed abnormalities on EEG. No unusual metabolites were malities of these disorders by replacing the supposed deficient found in plasma or urine, and metabolic testing was normal metabolites have not been successful in any case. except for persistent hypouricosuria. Investigation of purine This report describes four unrelated patients in whom and pyrimidine metabolism in cultured fibroblasts derived developmental delay, seizures, ataxia, recurrent infections, from these patients showed normal incorporation of purine speech deficit, and an unusual behavioral phenotype were bases into nucleotides but decreased incorporation of uridine. -

Identification of the Enzymatic Pathways of Nucleotide Metabolism in Human Lymphocytes and Leukemia Cells'
[CANCER RESEARCH 33, 94-103, January 1973] Identification of the Enzymatic Pathways of Nucleotide Metabolism in Human Lymphocytes and Leukemia Cells' E. M. Scholar and P. Calabresi Department of Medicine, The Roger Williams General Hospital, Providence, Rhode Island 02908, and The Division of Biological and Medical Sciences,Brown University, Providence,Rhode Island 02912 SUMMARY monocytes, and lymphocytes, it is difficult to know in what particular fraction an enzyme activity is present. It would therefore be advantageous to separate out the different Extracts of lymphocytes from normal donors and from components of the leukocyte fraction before investigating patients with chronic lymphocytic leukemia (CLL) and acute their enzymatic activities. All previously reported work on the lymphoblastic leukemia were examined for a variety of enzymes of purine nucleotide metabolism was done at best in enzymes with activity for purine nucleotide biosynthesis, the whole white blood cell fraction. Those enzymes found to interconversion, and catabolism as well as for a selected be present included adenine and guanine phosphoribosyltrans number of enzymes involved in pyrimidine nucleotide ferase (2, 32), PNPase2 (7), deoxyadenosine deaminase (7), metabolism. Lymphocytes from all three donor types (normal, ATPase (4), and inosine kinase (21). CLL, acute lymphocytic leukemia) contained the following A detailed knowledge of the enzymatic pathways of purine enzymatic activities: adenine and guanine phosphoribosyl and pyrimidine nucleotide metabolism in normal lymphocytes transferase , adenosine kinase , nucieoside diphosphate kinase, and leukemia cells is important in elucidating any biochemical adenylate kinase, guanylate kinase, cytidylate kinase, uridylate differences that may exist. Such differences may be exploited kinase, adenosine deaminase, purine nucleoside phosphorylase, in chemotherapy and in gaining and understanding of the and adenylate deaminase (with ATP). -

Source: the Arabidopsis Information Resource (TAIR);
Table S1 List of targeted loci and information about their function in Arabidopsis thaliana (source: The Arabidopsis Information Resource (TAIR); https://www.arabidopsis.org/tools/bulk/genes/index.jsp). Locus Gene Model Gene Model Description Gene Model Primary Gene Symbol All Gene Symbols Identifier Name Type AT1G78800 AT1G78800.1 UDP-Glycosyltransferase superfamily protein_coding protein;(source:Araport11) AT5G06830 AT5G06830.1 hypothetical protein;(source:Araport11) protein_coding AT2G31740 AT2G31740.1 S-adenosyl-L-methionine-dependent methyltransferases protein_coding superfamily protein;(source:Araport11) AT5G11960 AT5G11960.1 magnesium transporter, putative protein_coding (DUF803);(source:Araport11) AT4G00560 AT4G00560.4 NAD(P)-binding Rossmann-fold superfamily protein_coding protein;(source:Araport11) AT1G80510 AT1G80510.1 Encodes a close relative of the amino acid transporter ANT1 protein_coding (AT3G11900). AT2G21250 AT2G21250.1 NAD(P)-linked oxidoreductase superfamily protein_coding protein;(source:Araport11) AT5G04420 AT5G04420.1 Galactose oxidase/kelch repeat superfamily protein_coding protein;(source:Araport11) AT4G34910 AT4G34910.1 P-loop containing nucleoside triphosphate hydrolases protein_coding superfamily protein;(source:Araport11) AT5G66120 AT5G66120.2 3-dehydroquinate synthase;(source:Araport11) protein_coding AT1G45110 AT1G45110.1 Tetrapyrrole (Corrin/Porphyrin) protein_coding Methylase;(source:Araport11) AT1G67420 AT1G67420.2 Zn-dependent exopeptidases superfamily protein_coding protein;(source:Araport11) AT3G62370 -

O O2 Enzymes Available from Sigma Enzymes Available from Sigma
COO 2.7.1.15 Ribokinase OXIDOREDUCTASES CONH2 COO 2.7.1.16 Ribulokinase 1.1.1.1 Alcohol dehydrogenase BLOOD GROUP + O O + O O 1.1.1.3 Homoserine dehydrogenase HYALURONIC ACID DERMATAN ALGINATES O-ANTIGENS STARCH GLYCOGEN CH COO N COO 2.7.1.17 Xylulokinase P GLYCOPROTEINS SUBSTANCES 2 OH N + COO 1.1.1.8 Glycerol-3-phosphate dehydrogenase Ribose -O - P - O - P - O- Adenosine(P) Ribose - O - P - O - P - O -Adenosine NICOTINATE 2.7.1.19 Phosphoribulokinase GANGLIOSIDES PEPTIDO- CH OH CH OH N 1 + COO 1.1.1.9 D-Xylulose reductase 2 2 NH .2.1 2.7.1.24 Dephospho-CoA kinase O CHITIN CHONDROITIN PECTIN INULIN CELLULOSE O O NH O O O O Ribose- P 2.4 N N RP 1.1.1.10 l-Xylulose reductase MUCINS GLYCAN 6.3.5.1 2.7.7.18 2.7.1.25 Adenylylsulfate kinase CH2OH HO Indoleacetate Indoxyl + 1.1.1.14 l-Iditol dehydrogenase L O O O Desamino-NAD Nicotinate- Quinolinate- A 2.7.1.28 Triokinase O O 1.1.1.132 HO (Auxin) NAD(P) 6.3.1.5 2.4.2.19 1.1.1.19 Glucuronate reductase CHOH - 2.4.1.68 CH3 OH OH OH nucleotide 2.7.1.30 Glycerol kinase Y - COO nucleotide 2.7.1.31 Glycerate kinase 1.1.1.21 Aldehyde reductase AcNH CHOH COO 6.3.2.7-10 2.4.1.69 O 1.2.3.7 2.4.2.19 R OPPT OH OH + 1.1.1.22 UDPglucose dehydrogenase 2.4.99.7 HO O OPPU HO 2.7.1.32 Choline kinase S CH2OH 6.3.2.13 OH OPPU CH HO CH2CH(NH3)COO HO CH CH NH HO CH2CH2NHCOCH3 CH O CH CH NHCOCH COO 1.1.1.23 Histidinol dehydrogenase OPC 2.4.1.17 3 2.4.1.29 CH CHO 2 2 2 3 2 2 3 O 2.7.1.33 Pantothenate kinase CH3CH NHAC OH OH OH LACTOSE 2 COO 1.1.1.25 Shikimate dehydrogenase A HO HO OPPG CH OH 2.7.1.34 Pantetheine kinase UDP- TDP-Rhamnose 2 NH NH NH NH N M 2.7.1.36 Mevalonate kinase 1.1.1.27 Lactate dehydrogenase HO COO- GDP- 2.4.1.21 O NH NH 4.1.1.28 2.3.1.5 2.1.1.4 1.1.1.29 Glycerate dehydrogenase C UDP-N-Ac-Muramate Iduronate OH 2.4.1.1 2.4.1.11 HO 5-Hydroxy- 5-Hydroxytryptamine N-Acetyl-serotonin N-Acetyl-5-O-methyl-serotonin Quinolinate 2.7.1.39 Homoserine kinase Mannuronate CH3 etc. -
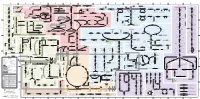
Fullsubwaymap221.Pdf
A B C D E F G H I J K L M Aldose reductase Sorbitol dehydrogenase Cholesterol synthesis Glycogen and galactose metabolism (branch point only) (debranching) glucose sorbitol fructose Aromatic amino acid metabolism Purine synthesis Pyrimidine synthesis glycogen (n+1) phenylacetate triiodothyronine (T3) dopaquinone melanin + + thyroxine (T4) (multiple steps) (many cycles) P 4' NADPH NADP NAD NADH acetyl-CoA x2 i Debranching enzyme 3' - α-1,4 bonds Aromatic amino acid 5-phosphoribosyl pyrophosphate (PRPP) HCO B 2' P 3 (branching) Glycogen 6 i (multiple steps) O H O (multiple steps) Tyrosinase O H O 1' 2 2 2 2 B 1 2 3 4 5 6 7 8 9 10 Pentose phosphate pathway Phenylalanine Tyrosine decarboxylase 6 Dopamine B Thiolase phosphorylase -5 -4 -3 -2 -1 0 1 2 3 4 Transaminase 6 glutamine 2 ATP (many cycles) Glycogen Glucose-6-phosphatase Dehydrogenase hydroxylase hydroxylase (DOPA decarboxylase) β-hydroxylase Methyltransferase 10 UDP 4:4 Transferase ATP glutathione (reduced) H O phenyllactate phenylpyruvate phenylalanine tyrosine dihydroxy- dopamine Glutamine PRPP CoA synthase Hexokinase or (in endoplasmic reticulum) 2 2 norepinephrine epinephrine glutamine Carbamoyl 9 1' phenylanine amidotransferase (GPAT) glutamate glycogen (n) Glutathione reductase Glutathione peroxidase + phosphate glycogen (n) -5 -4 -3 -2 -1 0 1 2 3 4 2' 3' 4' glucokinase 8 NADP NADPH glutamate α-ketoglutarate CO2 O H O Glycosyl (4:6) UDP-glucose ADP (L-DOPA) 2 2 synthetase II glutathione (oxidized) 2 H O α-ketoglutarate PPi glutamate acetoacetyl-CoA 7 1 Glucose -1,6-Glucosidase -

Myoadenylate Deaminase Deficiency. Functional and Metabolic Abnormalities Associated with Disruption of the Purine Nucleotide Cycle
Myoadenylate deaminase deficiency. Functional and metabolic abnormalities associated with disruption of the purine nucleotide cycle. R L Sabina, … , S DiMauro, E W Holmes J Clin Invest. 1984;73(3):720-730. https://doi.org/10.1172/JCI111265. Research Article To assess the role of the purine nucleotide cycle in human skeletal muscle function, we evaluated 10 patients with AMP deaminase deficiency (myoadenylate deaminase deficiency; MDD). 4 MDD and 19 non-MDD controls participated in an exercise protocol. The latter group was composed of a patient cohort (n = 8) exhibiting a constellation of symptoms similar to those of the MDD patients, i.e., postexertional aches, cramps, and pains; as well as a cohort of normal, unconditioned volunteers (n = 11). The individuals with MDD fatigued after performing only 28% as much work as their non-MDD counterparts. Muscle biopsies were obtained from the four MDD patients and the eight non-MDD patients at rest and following exercise to the point of fatigue. Creatine phosphate content fell to a comparable extent in the MDD (69%) and non-MDD (52%) patients at the onset of fatigue. Following exercise the 34% decrease in ATP content of muscle from the non-MDD subjects was significantly greater than the 6% decrease in ATP noted in muscle from the MDD patients (P = 0.048). Only one of four MDD patients had a measurable drop in ATP compared with seven of eight non- MDD patients. At end-exercise the muscle content of inosine 5'-monophosphate (IMP), a product of AMP deaminase, was 13-fold greater in the non-MDD patients than that observed in the MDD group […] Find the latest version: https://jci.me/111265/pdf Myoadenylate Deaminase Deficiency Functional and Metabolic Abnormalities Associated with Disruption of the Purine Nucleotide Cycle Richard L. -

1 Table S5. Genes Upregulated in Lectin+, KRT5-Gfphi Bcs
Rock et al., PNAS 2009 Table S5. Genes upregulated in lectin+, KRT5-GFPhi BCs versus non-BCs with p<0.001 and >21.5 fold change. Probe Symbol Name Fold 1435479_at --- --- 28.242 1436092_at --- Transcribed locus 13.519 1434848_at --- Transcribed locus 12.162 1438577_at --- Transcribed locus 8.387 1436651_at --- --- 6.605 1420194_at --- --- 4.812 1434209_at --- --- 3.904 1443669_at --- Transcribed locus 3.893 1439887_at --- 16 days embryo head cDNA, RIKEN full-length enriched library, clone:C130025B04 3.820 1441574_at --- Adult male diencephalon cDNA, RIKEN full-length enriched library, clone:9330190O21 3.767 1443529_at --- --- 3.683 1447314_at --- --- 3.680 1440485_at --- --- 3.596 1440371_at --- 2 days neonate sympathetic ganglion cDNA, RIKEN full-length enriched library, clone:7120457A19 3.595 1447988_at --- --- 3.554 1439990_at --- 12 days embryo spinal ganglion cDNA, RIKEN full-length enriched library, clone:D130065P14 3.544 1447204_at --- --- 3.539 1439769_at --- Transcribed locus 3.521 1439714_at --- --- 3.428 1458599_at --- --- 3.274 1445154_at --- --- 3.260 1445697_at --- Transcribed locus 3.253 1444425_at --- 0 day neonate kidney cDNA, RIKEN full-length enriched library, clone:D630017L16 3.233 1460556_at --- --- 3.163 1441341_at --- Transcribed locus 3.128 1446834_at --- Transcribed locus 3.113 1440592_at --- Transcribed locus 3.110 1456186_at --- --- 2.972 1447544_at --- Transcribed locus 2.862 1444479_at --- Transcribed locus 2.857 1436772_at --- Transcribed locus 2.851 1441368_at --- 12 days embryo embryonic body between diaphragm