Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Global Analysis Reveals the Complexity of the Human Glomerular Extracellular Matrix
Global analysis reveals the complexity of the human glomerular extracellular matrix Rachel Lennon,1,2 Adam Byron,1,* Jonathan D. Humphries,1 Michael J. Randles,1,2 Alex Carisey,1 Stephanie Murphy,1,2 David Knight,3 Paul E. Brenchley,2 Roy Zent,4,5 and Martin J. Humphries.1 1Wellcome Trust Centre for Cell-Matrix Research, Faculty of Life Sciences, University of Manchester, Manchester, UK; 2Faculty of Medical and Human Sciences, University of Manchester, Manchester, UK; 3Biological Mass Spectrometry Core Facility, Faculty of Life Sciences, University of Manchester, Manchester, UK; 4Division of Nephrology, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA; and 5Veterans Affairs Hospital, Nashville, TN, USA. *Present address: Edinburgh Cancer Research UK Centre, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK. Running title: Proteome of the glomerular matrix Word count: Abstract: 208, main text 2765 Corresponding author: Dr Rachel Lennon, Wellcome Trust Centre for Cell-Matrix Research, Michael Smith Building, University of Manchester, Manchester M13 9PT, UK. Phone: 0044 (0) 161 2755498. Fax: 0044 (0) 161 2755082. Email: [email protected] Abstract The glomerulus contains unique cellular and extracellular matrix (ECM) components, which are required for intact barrier function. Studies of the cellular components have helped to build understanding of glomerular disease; however, the full composition and regulation of glomerular ECM remains poorly understood. Here, we employed mass spectrometry–based proteomics of enriched ECM extracts for a global analysis of human glomerular ECM in vivo and identified a tissue-specific proteome of 144 structural and regulatory ECM proteins. This catalogue includes all previously identified glomerular components, plus many new and abundant components. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Matrix Gla Protein Species and Risk of Cardiovascular Events in Type 2 Diabetic Patients
Cardiovascular and Metabolic Risk ORIGINAL ARTICLE Matrix Gla Protein Species and Risk of Cardiovascular Events in Type 2 Diabetic Patients 1 3 GEERTJE W. DALMEIJER, PHD W.M. MONIQUE VERSCHUREN, PHD reduced coronary artery calcification and 1 3 YVONNE T. VAN DER SCHOUW, PHD JOLANDA M.A. BOER, PHD – 2 1 reduced risk of CVD (6 9). These effects ELKE J. MAGDELEYNS, BSC JOLINE W.J. BEULENS, PHD 2 are thought to be mediated by increased CEES VERMEER, PHD activation of MGP (10). MGP exists as various species, which OBJECTIVEd differ in their state of phosphorylation To investigate the relationship of circulating matrix Gla protein (MGP) species or carboxylation: phosphorylated, non- with incident cardiovascular disease (CVD) or coronary heart disease (CHD) in type 2 diabetic phosphorylated (desphospho-MGP patients. [dpMGP]), carboxylated (cMGP), or un- RESEARCH DESIGN AND METHODSdEPIC-NL is a prospective cohort study among carboxylated (ucMGP). Total uncarboxy- 40,011 Dutch men and women. At baseline (1993–1997), 518 participants were known to have lated MGP (t-ucMGP) is thought to be type 2 diabetes. MGP levels were measured by ELISA techniques in baseline plasma samples. The the sum of desphospho-uncarboxylated incidence of fatal and nonfatal CVD and CVD subtypesdCHD, peripheral arterial disease (PAD), MGP (dp-ucMGP) and phosphorylated- d heart failure, and stroke were obtained by linkage to national registers. Cox proportional uncarboxylated MGP (p-ucMGP) and hazard models were used to calculate hazard ratios (HRs), adjusted for sex, waist-to-hip ratio, mainly consists of p-ucMGP. physical activity, and history of CVD. Development of assays to measure RESULTSdDuring a median 11.2 years of follow-up, 160 cases of CVD were documented. -

Inactive Matrix Gla Protein Is a Novel Circulating Biomarker Predicting
www.nature.com/scientificreports OPEN Inactive matrix Gla protein is a novel circulating biomarker predicting retinal arteriolar Received: 5 June 2018 Accepted: 17 September 2018 narrowing in humans Published: xx xx xxxx Fang-Fei Wei1, Qi-Fang Huang1, Zhen-Yu Zhang1, Karel Van Keer2, Lutgarde Thijs1, Sander Trenson3, Wen-Yi Yang1, Nicholas Cauwenberghs 1, Blerim Mujaj1, Tatiana Kuznetsova1, Karel Allegaert4,5, Harry A. J. Struijker-Boudier6, Peter Verhamme7, Cees Vermeer8 & Jan A. Staessen 1,9 Active matrix Gla protein (MGP), a potent inhibitor of calcifcation in large arteries, protects against macrovascular complications. Recent studies suggested that active MGP helps maintaining the integrity of the renal and myocardial microcirculation, but its role in preserving the retinal microcirculation remains unknown. In 935 randomly recruited Flemish participants (mean age, 40.9 years; 50.3% women), we measured plasma desphospho-uncarboxylated MGP (dp–ucMGP), a marker of poor vitamin K status using an ELISA-based assay at baseline (1996–2010) and retinal microvascular diameters using IVAN software (Vasculomatic ala Nicola, version 1.1) including the central retinal arteriolar (CRAE) and venular (CRVE) equivalent and the arteriole-to-venule ratio (AVR) at follow-up (2008–2015). CRAE (P = 0.005) and AVR (P = 0.080) at follow-up decreased across tertiles of the dp–ucMGP distribution. In unadjusted models, for a doubling of dp–ucMGP at baseline, CRAE and AVR at follow-up respectively decreased by 1.40 µm (95% confdence interval [CI], 0.32 to 2.48; P = 0.011) and 0.006 (CI, 0.001 to 0.011; P = 0.016). In multivariable-adjusted models accounting for sex, baseline characteristics and follow-up duration, these estimates were −1.03 µm (CI, −1.96 to −0.11; P = 0.028) and −0.007 (CI, −0.011 to −0.002; P = 0.007). -
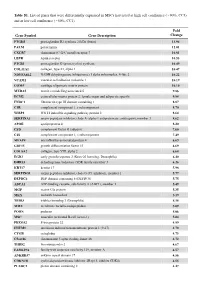
Table S1. List of Genes That Were Differentially Expressed in Mscs Harvested at High Cell Confluence (~90%, CC3) and at Low Cell Confluence (~50%, CC1)
Table S1. List of genes that were differentially expressed in MSCs harvested at high cell confluence (~90%, CC3) and at low cell confluence (~50%, CC1). Fold Gene Symbol Gene Description Change PTGDS prostaglandin D2 synthase 21kDa (brain) 13.94 PALM paralemmin 12.01 CXCR7 chemokine (C-X-C motif) receptor 7 10.81 LEPR leptin receptor 10.53 PTGIS prostaglandin I2 (prostacyclin) synthase 10.49 COL11A1 collagen, type XI, alpha 1 10.47 NDUFA4L2 NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 10.22 VCAM1 vascular cell adhesion molecule 1 10.19 COMP cartilage oligomeric matrix protein 10.10 MXRA5 matrix-remodelling associated 5 9.86 ECM2 extracellular matrix protein 2, female organ and adipocyte specific 9.84 FNDC1 fibronectin type III domain containing 1 8.87 C1R complement component 1, r subcomponent 8.70 WISP2 WNT1 inducible signaling pathway protein 2 8.64 SERPINA3 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 8.62 APOE apolipoprotein E 8.20 CFD complement factor D (adipsin) 7.60 C1S complement component 1, s subcomponent 7.49 MFAP4 microfibrillar-associated protein 4 6.69 GDF15 growth differentiation factor 15 6.69 COL8A2 collagen, type VIII, alpha 2 6.68 EGR2 early growth response 2 (Krox-20 homolog, Drosophila) 6.40 DHRS3 dehydrogenase/reductase (SDR family) member 3 6.26 KRT17 keratin 17 5.98 SERPING1 serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 5.77 DEPDC6 DEP domain containing 6 (DEPDC6) 5.75 ABCA1 ATP-binding cassette, sub-family A (ABC1), member 1 5.49 MGP matrix -

Cell-Deposited Matrix Improves Retinal Pigment Epithelium Survival on Aged Submacular Human Bruch’S Membrane
Retinal Cell Biology Cell-Deposited Matrix Improves Retinal Pigment Epithelium Survival on Aged Submacular Human Bruch’s Membrane Ilene K. Sugino,1 Vamsi K. Gullapalli,1 Qian Sun,1 Jianqiu Wang,1 Celia F. Nunes,1 Noounanong Cheewatrakoolpong,1 Adam C. Johnson,1 Benjamin C. Degner,1 Jianyuan Hua,1 Tong Liu,2 Wei Chen,2 Hong Li,2 and Marco A. Zarbin1 PURPOSE. To determine whether resurfacing submacular human most, as cell survival is the worst on submacular Bruch’s Bruch’s membrane with a cell-deposited extracellular matrix membrane in these eyes. (Invest Ophthalmol Vis Sci. 2011;52: (ECM) improves retinal pigment epithelial (RPE) survival. 1345–1358) DOI:10.1167/iovs.10-6112 METHODS. Bovine corneal endothelial (BCE) cells were seeded onto the inner collagenous layer of submacular Bruch’s mem- brane explants of human donor eyes to allow ECM deposition. here is no fully effective therapy for the late complications of age-related macular degeneration (AMD), the leading Control explants from fellow eyes were cultured in medium T cause of blindness in the United States. The prevalence of only. The deposited ECM was exposed by removing BCE. Fetal AMD-associated choroidal new vessels (CNVs) and/or geo- RPE cells were then cultured on these explants for 1, 14, or 21 graphic atrophy (GA) in the U.S. population 40 years and older days. The explants were analyzed quantitatively by light micros- is estimated to be 1.47%, with 1.75 million citizens having copy and scanning electron microscopy. Surviving RPE cells from advanced AMD, approximately 100,000 of whom are African explants cultured for 21 days were harvested to compare bestro- American.1 The prevalence of AMD increases dramatically with phin and RPE65 mRNA expression. -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. -

Identification of Potential Salivary Biomarker Panels for Oral
www.nature.com/scientificreports OPEN Identifcation of potential salivary biomarker panels for oral squamous cell carcinoma Anu Jain1, Chinmaya Narayana Kotimoole2, Sushmita Ghoshal3, Jaimanti Bakshi4, Aditi Chatterjee5,6, Thottethodi Subrahmanya Keshava Prasad2 & Arnab Pal1* Oral squamous cell carcinoma (OSCC) is one of the most prevalent cancers worldwide with the maximum number of incidences and deaths reported from India. One of the major causes of poor survival rate associated with OSCC has been attributed to late presentation due to non-availability of a biomarker. Identifcation of early diagnostic biomarker will help in reducing the disease morbidity and mortality. We validated 12 salivary proteins using targeted proteomics, identifed initially by relative quantifcation of salivary proteins on LC–MS, in OSCC patients and controls. Salivary AHSG (p = 0.0041**) and KRT6C (p = 0.002**) were upregulated in OSCC cases and AZGP1 (p ≤ 0.0001***), KLK1 (p = 0.006**) and BPIFB2 (p = 0.0061**) were downregulated. Regression modelling resulted in a signifcant risk prediction model (p < 0.0001***) consisting of AZGP1, AHSG and KRT6C for which ROC curve had AUC, sensitivity and specifcity of 82.4%, 78% and 73.5% respectively for all OSCC cases and 87.9%, 87.5% and 73.5% respectively for late stage (T3/T4) OSCC. AZGP1, AHSG, KRT6C and BPIFB2 together resulted in ROC curve (p < 0.0001***) with AUC, sensitivity and specifcity of 94%, 100% and 77.6% respectively for N0 cases while KRT6C and AZGP1 for N+ cases with ROC curve (p < 0.0001***) having AUC sensitivity and specifcity of 76.8%, 73% and 69.4%. Our data aids in the identifcation of biomarker panels for the diagnosis of OSCC cases with a diferential diagnosis between early and late- stage cases. -

Matrix Gla Protein Binds to Fibronectin and Enhances Cell Attachment and Spreading on Fibronectin
Hindawi Publishing Corporation International Journal of Cell Biology Volume 2014, Article ID 807013, 13 pages http://dx.doi.org/10.1155/2014/807013 Research Article Matrix Gla Protein Binds to Fibronectin and Enhances Cell Attachment and Spreading on Fibronectin Satoru Ken Nishimoto1,2,3 and Miyako Nishimoto1 1 Department of Microbiology, Immunology, and Biochemistry, University of Tennessee Health Science Center, Memphis, TN, USA 2 Department of Biomedical Engineering, University of Tennessee Health Science Center, Memphis, TN, USA 3 Department of Medical Education, University of Tennessee Health Science Center, Memphis, TN, USA Correspondence should be addressed to Satoru Ken Nishimoto; [email protected] Received 5 June 2014; Revised 7 August 2014; Accepted 8 August 2014; Published 21 August 2014 Academic Editor: Guido Tarone Copyright © 2014 S. K. Nishimoto and M. Nishimoto. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Matrix Gla protein (MGP) is a vitamin K-dependent, extracellular matrix protein. MGP is a calcification inhibitor of arteries and cartilage. However MGP is synthesized in many tissues and is especially enriched in embryonic tissues and in cancer cells. The presence of MGP in those instances does not correlate well with the calcification inhibitory role. This study explores a potential mechanism for MGP to bind to matrix proteins and alter cell matrix interactions. Methods.Todetermine whether MGP influences cell behavior through interaction with fibronectin, we studied MGP binding to fibronectin, theeffect of MGP on fibronectin mediated cell attachment and spreading and immunolocalized MGP and fibronectin. -

Emerging Roles of Matricellular Proteins in Systemic Sclerosis
International Journal of Molecular Sciences Review Emerging Roles of Matricellular Proteins in Systemic Sclerosis Daniel Feng 1,2 and Casimiro Gerarduzzi 1,2,3,* 1 Département de Pharmacologie et Physiologie, Faculté de Médecine, Université de Montréal, Montréal, QC H3T 1J4, Canada; [email protected] 2 Centre de recherche de l’Hôpital Maisonneuve-Rosemont, Faculté de Médecine, Centre affilié à l’Université de Montréal, Montréal, QC H1T 2M4, Canada 3 Département de Médecine, Faculté de Médecine, Université de Montréal, Montréal, QC H3T 1J4, Canada * Correspondence: [email protected]; Tel.: +1-514-252-3400 (ext. 2813) Received: 31 May 2020; Accepted: 13 June 2020; Published: 6 July 2020 Abstract: Systemic sclerosis is a rare chronic heterogenous disease that involves inflammation and vasculopathy, and converges in end-stage development of multisystem tissue fibrosis. The loss of tight spatial distribution and temporal expression of proteins in the extracellular matrix (ECM) leads to progressive organ stiffening, which is a hallmark of fibrotic disease. A group of nonstructural matrix proteins, known as matricellular proteins (MCPs) are implicated in dysregulated processes that drive fibrosis such as ECM remodeling and various cellular behaviors. Accordingly, MCPs have been described in the context of fibrosis in sclerosis (SSc) as predictive disease biomarkers and regulators of ECM synthesis, with promising therapeutic potential. In this present review, an informative summary of major MCPs is presented highlighting their clear correlations to SSc- fibrosis. Keywords: systemic sclerosis; fibrosis; extracellular matrix; matricellular proteins; myofibroblasts; biomarkers/therapeutics 1. Introduction Systemic sclerosis (SSc) is a rare idiopathic disease that presents as a trifecta of compounding chronic abnormalities driven by autoimmunity, vasculopathy, and systemic tissue fibrosis [1]. -

Substrate Softness Promotes Terminal Differentiation of Human
Substrate softness promotes terminal differentiation of human keratinocytes without altering their ability to proliferate back into a rigid environment Choua Ya, Mariana Carrancá, Dominique Sigaudo-Roussel, Philippe Faure, Berengere Fromy, Romain Debret To cite this version: Choua Ya, Mariana Carrancá, Dominique Sigaudo-Roussel, Philippe Faure, Berengere Fromy, et al.. Substrate softness promotes terminal differentiation of human keratinocytes without altering their ability to proliferate back into a rigid environment. Archives of Dermatological Research, Springer Verlag, 2019, 311 (10), pp.741-751. 10.1007/s00403-019-01962-5. hal-02322166 HAL Id: hal-02322166 https://hal.archives-ouvertes.fr/hal-02322166 Submitted on 7 Dec 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Substrate softness promotes terminal differentiation of human keratinocytes without altering their ability to proliferate back into a rigid environment Choua Ya1,2, Mariana Carrancá1, Dominique Sigaudo-Roussel1, Philippe Faure3, Bérengère Fromy1†, Romain Debret1†* 1 CNRS, University Lyon 1, UMR 5305, Laboratory of Tissue Biology and Therapeutic Engineering, IBCP, 7 Passage du Vercors, 69367 Lyon Cedex 07, France. 2 Isispharma, 29 Rue Maurice Flandin, 69003 Lyon, France. 3 Alpol Cosmétique, 140 Rue Pasteur, 01500 Château-Gaillard, France. -

Transcriptome Profiling Reveals the Complexity of Pirfenidone Effects in IPF
ERJ Express. Published on August 30, 2018 as doi: 10.1183/13993003.00564-2018 Early View Original article Transcriptome profiling reveals the complexity of pirfenidone effects in IPF Grazyna Kwapiszewska, Anna Gungl, Jochen Wilhelm, Leigh M. Marsh, Helene Thekkekara Puthenparampil, Katharina Sinn, Miroslava Didiasova, Walter Klepetko, Djuro Kosanovic, Ralph T. Schermuly, Lukasz Wujak, Benjamin Weiss, Liliana Schaefer, Marc Schneider, Michael Kreuter, Andrea Olschewski, Werner Seeger, Horst Olschewski, Malgorzata Wygrecka Please cite this article as: Kwapiszewska G, Gungl A, Wilhelm J, et al. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF. Eur Respir J 2018; in press (https://doi.org/10.1183/13993003.00564-2018). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2018 Copyright 2018 by the European Respiratory Society. Transcriptome profiling reveals the complexity of pirfenidone effects in IPF Grazyna Kwapiszewska1,2, Anna Gungl2, Jochen Wilhelm3†, Leigh M. Marsh1, Helene Thekkekara Puthenparampil1, Katharina Sinn4, Miroslava Didiasova5, Walter Klepetko4, Djuro Kosanovic3, Ralph T. Schermuly3†, Lukasz Wujak5, Benjamin Weiss6, Liliana Schaefer7, Marc Schneider8†, Michael Kreuter8†, Andrea Olschewski1,