Identification of Potential Salivary Biomarker Panels for Oral
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Global Analysis Reveals the Complexity of the Human Glomerular Extracellular Matrix
Global analysis reveals the complexity of the human glomerular extracellular matrix Rachel Lennon,1,2 Adam Byron,1,* Jonathan D. Humphries,1 Michael J. Randles,1,2 Alex Carisey,1 Stephanie Murphy,1,2 David Knight,3 Paul E. Brenchley,2 Roy Zent,4,5 and Martin J. Humphries.1 1Wellcome Trust Centre for Cell-Matrix Research, Faculty of Life Sciences, University of Manchester, Manchester, UK; 2Faculty of Medical and Human Sciences, University of Manchester, Manchester, UK; 3Biological Mass Spectrometry Core Facility, Faculty of Life Sciences, University of Manchester, Manchester, UK; 4Division of Nephrology, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA; and 5Veterans Affairs Hospital, Nashville, TN, USA. *Present address: Edinburgh Cancer Research UK Centre, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK. Running title: Proteome of the glomerular matrix Word count: Abstract: 208, main text 2765 Corresponding author: Dr Rachel Lennon, Wellcome Trust Centre for Cell-Matrix Research, Michael Smith Building, University of Manchester, Manchester M13 9PT, UK. Phone: 0044 (0) 161 2755498. Fax: 0044 (0) 161 2755082. Email: [email protected] Abstract The glomerulus contains unique cellular and extracellular matrix (ECM) components, which are required for intact barrier function. Studies of the cellular components have helped to build understanding of glomerular disease; however, the full composition and regulation of glomerular ECM remains poorly understood. Here, we employed mass spectrometry–based proteomics of enriched ECM extracts for a global analysis of human glomerular ECM in vivo and identified a tissue-specific proteome of 144 structural and regulatory ECM proteins. This catalogue includes all previously identified glomerular components, plus many new and abundant components. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Matrix Gla Protein Species and Risk of Cardiovascular Events in Type 2 Diabetic Patients
Cardiovascular and Metabolic Risk ORIGINAL ARTICLE Matrix Gla Protein Species and Risk of Cardiovascular Events in Type 2 Diabetic Patients 1 3 GEERTJE W. DALMEIJER, PHD W.M. MONIQUE VERSCHUREN, PHD reduced coronary artery calcification and 1 3 YVONNE T. VAN DER SCHOUW, PHD JOLANDA M.A. BOER, PHD – 2 1 reduced risk of CVD (6 9). These effects ELKE J. MAGDELEYNS, BSC JOLINE W.J. BEULENS, PHD 2 are thought to be mediated by increased CEES VERMEER, PHD activation of MGP (10). MGP exists as various species, which OBJECTIVEd differ in their state of phosphorylation To investigate the relationship of circulating matrix Gla protein (MGP) species or carboxylation: phosphorylated, non- with incident cardiovascular disease (CVD) or coronary heart disease (CHD) in type 2 diabetic phosphorylated (desphospho-MGP patients. [dpMGP]), carboxylated (cMGP), or un- RESEARCH DESIGN AND METHODSdEPIC-NL is a prospective cohort study among carboxylated (ucMGP). Total uncarboxy- 40,011 Dutch men and women. At baseline (1993–1997), 518 participants were known to have lated MGP (t-ucMGP) is thought to be type 2 diabetes. MGP levels were measured by ELISA techniques in baseline plasma samples. The the sum of desphospho-uncarboxylated incidence of fatal and nonfatal CVD and CVD subtypesdCHD, peripheral arterial disease (PAD), MGP (dp-ucMGP) and phosphorylated- d heart failure, and stroke were obtained by linkage to national registers. Cox proportional uncarboxylated MGP (p-ucMGP) and hazard models were used to calculate hazard ratios (HRs), adjusted for sex, waist-to-hip ratio, mainly consists of p-ucMGP. physical activity, and history of CVD. Development of assays to measure RESULTSdDuring a median 11.2 years of follow-up, 160 cases of CVD were documented. -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

Inactive Matrix Gla Protein Is a Novel Circulating Biomarker Predicting
www.nature.com/scientificreports OPEN Inactive matrix Gla protein is a novel circulating biomarker predicting retinal arteriolar Received: 5 June 2018 Accepted: 17 September 2018 narrowing in humans Published: xx xx xxxx Fang-Fei Wei1, Qi-Fang Huang1, Zhen-Yu Zhang1, Karel Van Keer2, Lutgarde Thijs1, Sander Trenson3, Wen-Yi Yang1, Nicholas Cauwenberghs 1, Blerim Mujaj1, Tatiana Kuznetsova1, Karel Allegaert4,5, Harry A. J. Struijker-Boudier6, Peter Verhamme7, Cees Vermeer8 & Jan A. Staessen 1,9 Active matrix Gla protein (MGP), a potent inhibitor of calcifcation in large arteries, protects against macrovascular complications. Recent studies suggested that active MGP helps maintaining the integrity of the renal and myocardial microcirculation, but its role in preserving the retinal microcirculation remains unknown. In 935 randomly recruited Flemish participants (mean age, 40.9 years; 50.3% women), we measured plasma desphospho-uncarboxylated MGP (dp–ucMGP), a marker of poor vitamin K status using an ELISA-based assay at baseline (1996–2010) and retinal microvascular diameters using IVAN software (Vasculomatic ala Nicola, version 1.1) including the central retinal arteriolar (CRAE) and venular (CRVE) equivalent and the arteriole-to-venule ratio (AVR) at follow-up (2008–2015). CRAE (P = 0.005) and AVR (P = 0.080) at follow-up decreased across tertiles of the dp–ucMGP distribution. In unadjusted models, for a doubling of dp–ucMGP at baseline, CRAE and AVR at follow-up respectively decreased by 1.40 µm (95% confdence interval [CI], 0.32 to 2.48; P = 0.011) and 0.006 (CI, 0.001 to 0.011; P = 0.016). In multivariable-adjusted models accounting for sex, baseline characteristics and follow-up duration, these estimates were −1.03 µm (CI, −1.96 to −0.11; P = 0.028) and −0.007 (CI, −0.011 to −0.002; P = 0.007). -

Proteomic Expression Profile in Human Temporomandibular Joint
diagnostics Article Proteomic Expression Profile in Human Temporomandibular Joint Dysfunction Andrea Duarte Doetzer 1,*, Roberto Hirochi Herai 1 , Marília Afonso Rabelo Buzalaf 2 and Paula Cristina Trevilatto 1 1 Graduate Program in Health Sciences, School of Medicine, Pontifícia Universidade Católica do Paraná (PUCPR), Curitiba 80215-901, Brazil; [email protected] (R.H.H.); [email protected] (P.C.T.) 2 Department of Biological Sciences, Bauru School of Dentistry, University of São Paulo, Bauru 17012-901, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-41-991-864-747 Abstract: Temporomandibular joint dysfunction (TMD) is a multifactorial condition that impairs human’s health and quality of life. Its etiology is still a challenge due to its complex development and the great number of different conditions it comprises. One of the most common forms of TMD is anterior disc displacement without reduction (DDWoR) and other TMDs with distinct origins are condylar hyperplasia (CH) and mandibular dislocation (MD). Thus, the aim of this study is to identify the protein expression profile of synovial fluid and the temporomandibular joint disc of patients diagnosed with DDWoR, CH and MD. Synovial fluid and a fraction of the temporomandibular joint disc were collected from nine patients diagnosed with DDWoR (n = 3), CH (n = 4) and MD (n = 2). Samples were subjected to label-free nLC-MS/MS for proteomic data extraction, and then bioinformatics analysis were conducted for protein identification and functional annotation. The three Citation: Doetzer, A.D.; Herai, R.H.; TMD conditions showed different protein expression profiles, and novel proteins were identified Buzalaf, M.A.R.; Trevilatto, P.C. -

Supporting Information for Proteomics DOI 10.1002/Pmic.200401336
Supporting Information for Proteomics DOI 10.1002/pmic.200401336 Ilan Beer, Eilon Barnea, Arie Admon Centralized data analysis of a large interlaboratory proteomics project-A feasibility study ª 2005 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim www.proteomics-journal.de Number Number Amino Number of high IPI of acid Protein name of labs quality peptides coverage spectra IPI00022229.1 236 8 3490 59 Apolipoprotein B-100 precursor IPI00164623.2 151 8 12671 82 IPI00032257.1 150 4 12628 83 complement component 3 precursor IPI00022434.1 110 8 13599 94 Serum albumin precursor Complement C4 precursor [Contains: C4A IPI00032258.2 99 8 5449 58 anaphylatox IPI00022463.1 94 8 9905 82 Serotransferrin precursor IPI00032256.1 92 8 15676 67 Alpha-2-macroglobulin precursor IPI00328461.1 84 8 2103 51 Hypothetical protein DKFZp686H112 Complement C5 precursor [Contains: C5a IPI00032291.1 74 6 591 50 anaphylatox IPI00179357.1 68 8 20 2 Titin IPI00017601.1 66 8 3910 65 Ceruloplasmin precursor IPI00029739.1 65 8 2265 57 Splice isoform 1 of P08603 IPI00182635.1 65 8 1358 46 IPI00220327.1 56 7 4729 61 keratin 1 IPI00021841.1 51 8 7728 91 Apolipoprotein A-I precursor IPI00019580.1 51 6 701 67 Plasminogen precursor IPI00025426.1 48 4 3184 38 Pregnancy zone protein precursor IPI00021304.1 48 4 2241 68 Keratin, type II cytoskeletal 2 epidermal IPI00013933.1 48 4 59 20 Splice isoform DPI of P15924 IPI00294193.1 46 8 1916 56 Splice isoform 1 of Q14624 IPI00021885.1 45 7 1811 45 Splice isoform Alpha-E of P02671 IPI00305457.1 44 8 5917 76 Alpha-1-antitrypsin precursor -

Genome Wide Assessment of Early Osseointegration in Implant-Adherent Cells
Genome Wide assessment of Early Osseointegration in Implant-Adherent Cells Ghadeer N. Thalji A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the School of Dentistry (Oral Biology) Chapel Hill 2012 Approved by Lyndon F.Cooper, DDS, PhD Eric T. Everett, PhD Gustavo Mendonça, DDS, MS, PhD Salvador Nares DDS, PhD Clark M. Stanford, DDS, PhD © 2012 Ghadeer N. Thalji ALL RIGHTS RESERVED ii ABSTRACT GHADEER N. THALJI: Genome wide assessment of early osseointegration in implant-adherent cells (Under the direction of Professor Lyndon F. Cooper) Objectives: To determine the molecular processes involved in osseointegration. Materials and methods: A structured literature review concerning in vitro and in vivo molecular assessment of osseointegration was performed. A rat and a human model were then used to identify the early molecular processes involved in osseointegration associated with a micro roughened and nanosurface superimposed featured implants. In the rat model, 32 titanium implants with surface topographies exhibiting a micro roughened (AT-II) and nanosurface superimposed featured implants (AT-I) were placed in the tibiae of 8 rats and subsequently harvested at 2 and 4 days after placement. Whereas in the human model, four titanium mini-implants with either a moderately roughened surface (TiOblast) or super-imposed nanoscale topography (Osseospeed) were placed in edentulous sites of eleven systemically healthy subjects and subsequently removed after 3 and 7 days. Total RNA was isolated from cells adherent to retrieved implants. A whole genome microarray using the Affymetrix 1.1 ST Array platform was used to describe the gene expression profiles that were differentially regulated by the implant surfaces. -

Análisis De Los Perfiles De
“ANÁLISIS DE LOS PERFILES DE EXPRESIÓN DE GENES RELACIONADOS CON VÍAS DE PROLIFERACIÓN CELULAR EN FIBROBLASTOS DE CALVARIA DE UNA MUESTRA DE PACIENTES COLOMBIANOS CON SÍNDROME DE APERT POSTERIOR A LA DEGRADACIÓN HEPARÁN SULFATO” Diana Alexandra Ramírez Montaño Universidad Nacional de Colombia Facultad de Medicina Maestría en Genética Humana Bogotá, Colombia 2016 “ANÁLISIS DE LOS PERFILES DE EXPRESIÓN DE GENES RELACIONADOS CON VÍAS DE PROLIFERACIÓN CELULAR EN FIBROBLASTOS DE CALVARIA DE UNA MUESTRA DE PACIENTES COLOMBIANOS CON SÍNDROME DE APERT POSTERIOR A LA DEGRADACIÓN HEPARÁN SULFATO” Diana Alexandra Ramírez Montaño Tesis presentada como requisito parcial para optar al título de: Magister en Genética Humana Director: Harvy Mauricio Velasco Parra, MD Especialista en Genética Médica MsC Ciencias Biológicas Línea de Investigación: Anomalías congénitas Grupo de investigación: Genética Clínica Universidad Nacional de Colombia Facultad de Medicina Maestría en Genética Humana Bogotá, Colombia 2016 VII A mi madre, por ella y para ella. X Agradecimientos Agradezco inicialmente a Dios por cada paso que me ha permitido dar en mi vida.Gracias al Dr. Harvy Velasco, por convertirse desde hace varios años en mi mentor y guía en el camino de la investigación, por la confianza en mis capacidades académicas y por la paciencia durante todos estos años. A la Dra. Lilian Torres y al equipo de la Fundación Universitaria en Ciencias de la Salud por su apoyo para la realización de este proyecto. Así mismo, al Dr. Ángel Carracedo y su equipo de la Fundación Pública de Medicina Xenómica en la Universidad de Santiago de Compostela por sus aportes en el análisis de datos que permitieron la interpretación y aprovechamiento de los datos obtenidos. -

UCLA Electronic Theses and Dissertations
UCLA UCLA Electronic Theses and Dissertations Title Proteomic Analysis of Cancer Cell Metabolism Permalink https://escholarship.org/uc/item/8t36w919 Author Chai, Yang Publication Date 2013 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA Los Angeles Proteomic Analysis of Cancer Cell Metabolism A thesis submitted in partial satisfaction of the requirements of the degree Master of Science in Oral Biology by Yang Chai 2013 ABSTRACT OF THESIS Proteomic Analysis of Cancer Cell Metabolism by Yang Chai Master of Science in Oral Biology University of California, Los Angeles, 2013 Professor Shen Hu, Chair Tumor cells can adopt alternative metabolic pathways during oncogenesis. This is an event characterized by an enhanced utilization of glucose for rapid synthesis of macromolecules such as nucleotides, lipids and proteins. This phenomenon was also known as the ‘Warburg effect’, distinguished by a shift from oxidative phosphorylation to increased aerobic glycolysis in many types of cancer cells. Increased aerobic glycolysis was also indicated with enhanced lactate production and glutamine consumption, and has been suggested to confer growth advantage for proliferating cells during oncogenic transformation. Development of a tracer-based ii methodology to determine de novo protein synthesis by tracing metabolic pathways from nutrient utilization may certainly enhance current understanding of nutrient gene interaction in cancer cells. We hypothesized that the metabolic phenotype of cancer cells as characterized by nutrient utilization for protein synthesis is significantly altered during oncogenesis, and 13C stable isotope tracers may incorporate 13C into non-essential amino acids of protein peptides during de novo protein synthesis to reflect the underlying mechanisms in cancer cell metabolism. -
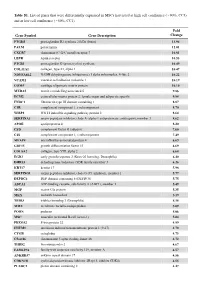
Table S1. List of Genes That Were Differentially Expressed in Mscs Harvested at High Cell Confluence (~90%, CC3) and at Low Cell Confluence (~50%, CC1)
Table S1. List of genes that were differentially expressed in MSCs harvested at high cell confluence (~90%, CC3) and at low cell confluence (~50%, CC1). Fold Gene Symbol Gene Description Change PTGDS prostaglandin D2 synthase 21kDa (brain) 13.94 PALM paralemmin 12.01 CXCR7 chemokine (C-X-C motif) receptor 7 10.81 LEPR leptin receptor 10.53 PTGIS prostaglandin I2 (prostacyclin) synthase 10.49 COL11A1 collagen, type XI, alpha 1 10.47 NDUFA4L2 NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 10.22 VCAM1 vascular cell adhesion molecule 1 10.19 COMP cartilage oligomeric matrix protein 10.10 MXRA5 matrix-remodelling associated 5 9.86 ECM2 extracellular matrix protein 2, female organ and adipocyte specific 9.84 FNDC1 fibronectin type III domain containing 1 8.87 C1R complement component 1, r subcomponent 8.70 WISP2 WNT1 inducible signaling pathway protein 2 8.64 SERPINA3 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 8.62 APOE apolipoprotein E 8.20 CFD complement factor D (adipsin) 7.60 C1S complement component 1, s subcomponent 7.49 MFAP4 microfibrillar-associated protein 4 6.69 GDF15 growth differentiation factor 15 6.69 COL8A2 collagen, type VIII, alpha 2 6.68 EGR2 early growth response 2 (Krox-20 homolog, Drosophila) 6.40 DHRS3 dehydrogenase/reductase (SDR family) member 3 6.26 KRT17 keratin 17 5.98 SERPING1 serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 5.77 DEPDC6 DEP domain containing 6 (DEPDC6) 5.75 ABCA1 ATP-binding cassette, sub-family A (ABC1), member 1 5.49 MGP matrix -

Cell-Deposited Matrix Improves Retinal Pigment Epithelium Survival on Aged Submacular Human Bruch’S Membrane
Retinal Cell Biology Cell-Deposited Matrix Improves Retinal Pigment Epithelium Survival on Aged Submacular Human Bruch’s Membrane Ilene K. Sugino,1 Vamsi K. Gullapalli,1 Qian Sun,1 Jianqiu Wang,1 Celia F. Nunes,1 Noounanong Cheewatrakoolpong,1 Adam C. Johnson,1 Benjamin C. Degner,1 Jianyuan Hua,1 Tong Liu,2 Wei Chen,2 Hong Li,2 and Marco A. Zarbin1 PURPOSE. To determine whether resurfacing submacular human most, as cell survival is the worst on submacular Bruch’s Bruch’s membrane with a cell-deposited extracellular matrix membrane in these eyes. (Invest Ophthalmol Vis Sci. 2011;52: (ECM) improves retinal pigment epithelial (RPE) survival. 1345–1358) DOI:10.1167/iovs.10-6112 METHODS. Bovine corneal endothelial (BCE) cells were seeded onto the inner collagenous layer of submacular Bruch’s mem- brane explants of human donor eyes to allow ECM deposition. here is no fully effective therapy for the late complications of age-related macular degeneration (AMD), the leading Control explants from fellow eyes were cultured in medium T cause of blindness in the United States. The prevalence of only. The deposited ECM was exposed by removing BCE. Fetal AMD-associated choroidal new vessels (CNVs) and/or geo- RPE cells were then cultured on these explants for 1, 14, or 21 graphic atrophy (GA) in the U.S. population 40 years and older days. The explants were analyzed quantitatively by light micros- is estimated to be 1.47%, with 1.75 million citizens having copy and scanning electron microscopy. Surviving RPE cells from advanced AMD, approximately 100,000 of whom are African explants cultured for 21 days were harvested to compare bestro- American.1 The prevalence of AMD increases dramatically with phin and RPE65 mRNA expression.