Supplemental Table 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Decreased Expression of Profilin 2 in Oral Squamous Cell Carcinoma and Its Clinicopathological Implications
ONCOLOGY REPORTS 26: 813-823, 2011 Decreased expression of profilin 2 in oral squamous cell carcinoma and its clinicopathological implications C.Y. MA1,2, C.P. ZHANG1,2, L.P. ZHONG1,2, H.Y. PAN1,2, W.T. CHEN1,2, L.Z. WANG3, O.W. ANDREW4, T. JI1 and W. HAN1,2 1Department of Oral and Maxillofacial Surgery, Ninth People's Hospital, College of Stomatology; 2Shanghai Key Laboratory of Stomatology and Shanghai Research Institute of Stomatology; 3Department of Oral Pathology, Ninth People's Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, P.R. China; 4Department of Oral and Maxillofacial Surgery, Faculty of Dentistry, National University of Singapore, Singapore 119074, Singapore Received February 8, 2011; Accepted April 11, 2011 DOI: 10.3892/or.2011.1365 Abstract. Profilins are small proteins essential for many clinical and pathological significance. In conclusion, PFN2 normal cellular dynamics and constitute one of the crucial can be utilized as both a potential suppressor marker and a components of actin-based cellular motility. Several recent prognostic protein for OSCC. The function of PFN2 may be to studies have implicated a role for the profilin (PFN) family in regulate the N-WASP/Arp2/3 signaling pathway. cancer pathogenesis and progression. However, their expression and promising functions are largely unknown in oral squamous Introduction cell carcinoma (OSCC). In this study, we analyzed the correlation between PFN1 and PFN2 expression in vitro and Oral squamous cell carcinoma (OSCC) is a significant public in vivo. The protein expression levels were roughly compared health problem with >300,000 new cases being diagnosed between cell lines (HIOEC, HB96) with the employment of annually worldwide (1). -

Associated Palmoplantar Keratoderma
DR ABIGAIL ZIEMAN (Orcid ID : 0000-0001-8236-207X) Article type : Review Article Pathophysiology of pachyonychia congenita-associated palmoplantar keratoderma: New insight into skin epithelial homeostasis and avenues for treatment Authors: A. G. Zieman1 and P. A. Coulombe1,2 # Affiliations: 1Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, MI 48109, USA; 2Department of Dermatology, University of Michigan Medical School, Ann Arbor, MI 48109, USA #Corresponding author: Pierre A. Coulombe, PhD, 3071 Biomedical Sciences Research Building, 109 Zina Pitcher Place, Ann Arbor, MI 48109, USA. Tel: 734-615-7509. Email: [email protected]. Funding Sources: These studies were supported by grant AR044232 issued to P.A.C. from the National Institute of Arthritis, Musculoskeletal and Skin Disease (NIAMS). A.G.Z. received support from grant T32 CA009110 from the National Cancer Institute. Author Manuscript This is the author manuscript accepted for publication and has undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/BJD.18033 This article is protected by copyright. All rights reserved Conflict of interest disclosures: None declared. Bulleted statements: What’s already known about this topic? Pachyonychia congenita is a rare genodermatosis caused by mutations in KRT6A, KRT6B, KRT6C, KRT16, KRT17, which are normally expressed in skin appendages and induced following injury. Individuals with PC present with multiple clinical symptoms that usually include thickened and dystrophic nails, palmoplantar keratoderma (PPK), glandular cysts, and oral leukokeratosis. -

Genome-Wide Analyses Identify KIF5A As a Novel ALS Gene
This is a repository copy of Genome-wide Analyses Identify KIF5A as a Novel ALS Gene.. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/129590/ Version: Accepted Version Article: Nicolas, A, Kenna, KP, Renton, AE et al. (210 more authors) (2018) Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron, 97 (6). 1268-1283.e6. https://doi.org/10.1016/j.neuron.2018.02.027 Reuse This article is distributed under the terms of the Creative Commons Attribution-NonCommercial-NoDerivs (CC BY-NC-ND) licence. This licence only allows you to download this work and share it with others as long as you credit the authors, but you can’t change the article in any way or use it commercially. More information and the full terms of the licence here: https://creativecommons.org/licenses/ Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Genome-wide Analyses Identify KIF5A as a Novel ALS Gene Aude Nicolas1,2, Kevin P. Kenna2,3, Alan E. Renton2,4,5, Nicola Ticozzi2,6,7, Faraz Faghri2,8,9, Ruth Chia1,2, Janice A. Dominov10, Brendan J. Kenna3, Mike A. Nalls8,11, Pamela Keagle3, Alberto M. Rivera1, Wouter van Rheenen12, Natalie A. Murphy1, Joke J.F.A. van Vugt13, Joshua T. Geiger14, Rick A. Van der Spek13, Hannah A. Pliner1, Shankaracharya3, Bradley N. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -
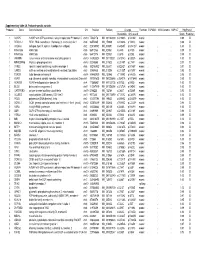
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Keratin 9 Point Mutation in the Pedigree of Epidermolytic Hereditary Palmoplantar Keratoderma Perturbs Keratin Intermediate Filament Network Formation
FEBS 17004 FEBS Letters 386 (1996) 149-155 Keratin 9 point mutation in the pedigree of epidermolytic hereditary palmoplantar keratoderma perturbs keratin intermediate filament network formation Setsu Kobayashi, Toshihiro Tanaka*, Norihisa Matsuyoshi, Sadao Imamura Department of Dermatology, Graduate School of Medicine, Kyoto University, Kyoto, 606 Japan Received 12 January 1996; revised version received 4 April 1996 Abstract Keratins form an intracellular keratin filament net- point mutations in the K9 gene in EHPPK [4-8] but none work in keratinocytes. Point mutations in the epidermal keratins showed a function assay with these mutations. Here, we pro- could lead to the disruption of keratin filament formation, vide the first demonstration that the point mutation found in developing skin diseases such as epidermolytic hereditary a pedigree of EHPPK has a dominant-negative effect on the palmoplantar keratoderma (EHPPK). We found a G to A assembly of keratin intermediate filaments in the cells. transition in keratin 9 (K9) cDNA, resulting in the substitution of glutamine for arginine at 162, in all patients of a pedigree of 2. Materials and methods EHPPK. Transfection into MDCK cells and DJM-1 cells revealed that the plasmid CMX vector containing normal keratin 2.1. PCR and DNA sequence 9 cDNA showed normal keratin network formation, whereas the Genomic DNA was extracted and purified from blood or biopsy vector with a G to A point mutated keratin 9 cDNA showed specimens from the patients. The primers were designed at nucleotide disrupted keratin filaments with droplet formation in the cells. 263 282 and 664-683 based on the K9 cDNA sequence [9]. -

CDH12 Cadherin 12, Type 2 N-Cadherin 2 RPL5 Ribosomal
5 6 6 5 . 4 2 1 1 1 2 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 A A A A A A A A A A A A A A A A A A A A C C C C C C C C C C C C C C C C C C C C R R R R R R R R R R R R R R R R R R R R B , B B B B B B B B B B B B B B B B B B B , 9 , , , , 4 , , 3 0 , , , , , , , , 6 2 , , 5 , 0 8 6 4 , 7 5 7 0 2 8 9 1 3 3 3 1 1 7 5 0 4 1 4 0 7 1 0 2 0 6 7 8 0 2 5 7 8 0 3 8 5 4 9 0 1 0 8 8 3 5 6 7 4 7 9 5 2 1 1 8 2 2 1 7 9 6 2 1 7 1 1 0 4 5 3 5 8 9 1 0 0 4 2 5 0 8 1 4 1 6 9 0 0 6 3 6 9 1 0 9 0 3 8 1 3 5 6 3 6 0 4 2 6 1 0 1 2 1 9 9 7 9 5 7 1 5 8 9 8 8 2 1 9 9 1 1 1 9 6 9 8 9 7 8 4 5 8 8 6 4 8 1 1 2 8 6 2 7 9 8 3 5 4 3 2 1 7 9 5 3 1 3 2 1 2 9 5 1 1 1 1 1 1 5 9 5 3 2 6 3 4 1 3 1 1 4 1 4 1 7 1 3 4 3 2 7 6 4 2 7 2 1 2 1 5 1 6 3 5 6 1 3 6 4 7 1 6 5 1 1 4 1 6 1 7 6 4 7 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

Comparative Genomics Analyses of Alpha-Keratins Reveal Insights Into
Sun et al. Frontiers in Zoology (2017) 14:41 DOI 10.1186/s12983-017-0225-x RESEARCH Open Access Comparative genomics analyses of alpha- keratins reveal insights into evolutionary adaptation of marine mammals Xiaohui Sun, Zepeng Zhang, Yingying Sun, Jing Li, Shixia Xu* and Guang Yang* Abstract Background: Diversity of hair in marine mammals was suggested as an evolutionary innovation to adapt aquatic environment, yet its genetic basis remained poorly explored. We scanned α-keratin genes, one major structural components of hair, in 16 genomes of mammalian species, including seven cetaceans, two pinnipeds, polar bear, manatee and five terrestrial species. Results: Extensive gene loss and high pseudogenization rate of α-keratin genes were identified in cetaceans when compared to terrestrial artiodactylans (average number of α-keratins 37.29 vs. 58.33; pseudogenization rate 29.89% vs. 8.00%), especially of hair follicle-specific keratin genes (average pseudogenization rate in cetaceans of 43.88% relative to 3.80% artiodactylian average). Compared to toothed whale, the much more number of intact functional α-keratin genes was examined in the baleen whale that had specific keratinized baleen. In contrast, the number of keratin genes in pinnipeds, polar bear and manatee were comparable to those of their respective terrestrial relatives. Additionally, four keratin genes (K39, K9, K42, and K74) were found to be pseudogenes or lost uniquely in cetaceans and manatees. Conclusions: Species-specific evolution of α-keratin gene family identified in the marine mammals might be responsible for their different hair characteristics. Increased gene loss and pseudogenization rate identified in cetacean lineages was likely to contribute to hair-less phenotype to adaptation for complete aquatic environment. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Exploring Profilin
RESEARCH HIGHLIGHTS NEUROTRANSMISSION Exploring profilin DOI: 10.1038/nrn2182 Surprisingly, they found that Pfn2–/– mice, pointing towards a Pfn2–/– mice displayed normal higher vesicle release probability. development and brain anatomy. Immunoprecipitation and EM studies Learning and memory, long-term also indicated that the number of potentiation (LTP) and long-term docked vesicles is approximately 25% depression (LTD) were not impaired higher in Pfn2–/– mice. in Pfn2–/– mice when compared with How does profilin 2 alter vesicle control mice. These results suggest release? When neurotransmitter that profilin 2 is not required for secretion was stimulated in cortical development, synaptic plasticity or synaptosomes from control mice, learning and memory. F-actin levels were significantly The authors noticed, however, increased. This did not occur in that the Pfn2–/– mice behaved synaptosome preparations from differently to control mice. They Pfn2–/– mice. In addition, profilin 2, were hyperactive, and they showed but not profilin 1, was shown to be elevated exploratory and novelty- tightly associated with the WAVE- Profilins are regulators of actin seeking behaviour when placed into complex, a protein complex that is polymerization. Profilin 2 is a new environment. This included implicated in actin dynamics and expressed mainly in neurons, but increased locomotor activity, wall is abundant in synaptosomal prepa- its specific function is unknown. A rearings and rearings in the centre of rations. The authors therefore study by Boyl et al. provides evidence the experimental area. Conversely, suggest that WAVE and profilin 2 that profilin 2 is required for activ- in the familiar environment of their might cooperate to restrict synaptic ity-stimulated actin polymerization home cage, they showed reduced vesicle release. -

Profilin and Formin Constitute a Pacemaker System for Robust Actin
RESEARCH ARTICLE Profilin and formin constitute a pacemaker system for robust actin filament growth Johanna Funk1, Felipe Merino2, Larisa Venkova3, Lina Heydenreich4, Jan Kierfeld4, Pablo Vargas3, Stefan Raunser2, Matthieu Piel3, Peter Bieling1* 1Department of Systemic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund, Germany; 2Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund, Germany; 3Institut Curie UMR144 CNRS, Paris, France; 4Physics Department, TU Dortmund University, Dortmund, Germany Abstract The actin cytoskeleton drives many essential biological processes, from cell morphogenesis to motility. Assembly of functional actin networks requires control over the speed at which actin filaments grow. How this can be achieved at the high and variable levels of soluble actin subunits found in cells is unclear. Here we reconstitute assembly of mammalian, non-muscle actin filaments from physiological concentrations of profilin-actin. We discover that under these conditions, filament growth is limited by profilin dissociating from the filament end and the speed of elongation becomes insensitive to the concentration of soluble subunits. Profilin release can be directly promoted by formin actin polymerases even at saturating profilin-actin concentrations. We demonstrate that mammalian cells indeed operate at the limit to actin filament growth imposed by profilin and formins. Our results reveal how synergy between profilin and formins generates robust filament growth rates that are resilient to changes in the soluble subunit concentration. DOI: https://doi.org/10.7554/eLife.50963.001 *For correspondence: peter.bieling@mpi-dortmund. mpg.de Introduction Competing interests: The Eukaryotic cells move, change their shape and organize their interior through dynamic actin net- authors declare that no works.