Saccostrea Glomerata)CO2 Stress Response Nicole G
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diseases Affecting Finfish
Diseases Affecting Finfish Legislation Ireland's Exotic / Disease Name Acronym Health Susceptible Species Vector Species Non-Exotic Listed National Status Disease Measures Bighead carp (Aristichthys nobilis), goldfish (Carassius auratus), crucian carp (C. carassius), Epizootic Declared Rainbow trout (Oncorhynchus mykiss), redfin common carp and koi carp (Cyprinus carpio), silver carp (Hypophtalmichthys molitrix), Haematopoietic EHN Exotic * Disease-Free perch (Percha fluviatilis) Chub (Leuciscus spp), Roach (Rutilus rutilus), Rudd (Scardinius erythrophthalmus), tench Necrosis (Tinca tinca) Beluga (Huso huso), Danube sturgeon (Acipenser gueldenstaedtii), Sterlet sturgeon (Acipenser ruthenus), Starry sturgeon (Acipenser stellatus), Sturgeon (Acipenser sturio), Siberian Sturgeon (Acipenser Baerii), Bighead carp (Aristichthys nobilis), goldfish (Carassius auratus), Crucian carp (C. carassius), common carp and koi carp (Cyprinus carpio), silver carp (Hypophtalmichthys molitrix), Chub (Leuciscus spp), Roach (Rutilus rutilus), Rudd (Scardinius erythrophthalmus), tench (Tinca tinca) Herring (Cupea spp.), whitefish (Coregonus sp.), North African catfish (Clarias gariepinus), Northern pike (Esox lucius) Catfish (Ictalurus pike (Esox Lucius), haddock (Gadus aeglefinus), spp.), Black bullhead (Ameiurus melas), Channel catfish (Ictalurus punctatus), Pangas Pacific cod (G. macrocephalus), Atlantic cod (G. catfish (Pangasius pangasius), Pike perch (Sander lucioperca), Wels catfish (Silurus glanis) morhua), Pacific salmon (Onchorhynchus spp.), Viral -

An Assessment of Potential Heavy Metal Contaminants in Bivalve Shellfish from Aquaculture Zones Along the Coast of New South Wales, Australia
PEER-REVIEWED ARTICLE Hazel Farrell,* Phil Baker, Grant Webster, Food Protection Trends, Vol 38, No. 1, p. 18–26 Copyright© 2018, International Association for Food Protection Edward Jansson, Elizabeth Szabo and 6200 Aurora Ave., Suite 200W, Des Moines, IA 50322-2864 Anthony Zammit NSW Food Authority, 6 Ave. of the Americas, Newington NSW 2127, Australia An Assessment of Potential Heavy Metal Contaminants in Bivalve Shellfish from Aquaculture Zones along the Coast of New South Wales, Australia ABSTRACT INTRODUCTION Evaluation of shellfish aquaculture for potential contam- Certain elements are essential in human physiology; inants is essential for consumer confidence and safety. however, an incorrect balance or excess of certain elements Every three years, between 1999 and 2014, bivalve in the diet can result in negative health effects. Heavy metals shellfish from aquaculture zones in up to 31 estuaries are of particular concern because of their ability to persist across 2,000 km of Australia’s east coast were tested and accumulate in the environment. While heavy metals for cadmium, copper, lead, mercury, selenium and zinc. can occur naturally in the environment, human activities Inorganic arsenic was included in the analyses in 2002, and run-off from urban and agricultural land use may and total arsenic was used as a screen for the inorganic increase their concentrations (6, 29). This is particularly form in subsequent years. Concentrations of inorganic important when considering the growing demands on arsenic, cadmium, lead and mercury were low and did not coastal resources due to increasing populations (3) and the exceed maximum limits mandated in the Australia New ability of filter feeding bivalve shellfish to bio-accumulate Zealand Food Standards Code. -
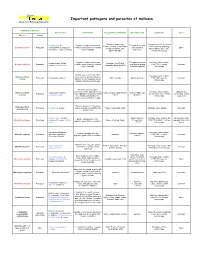
Pathogens Table 2006
Important pathogens and parasites of molluscs Genetic and Pathology Laboratory Pathogen or parasite Host species Host impact Geographical distribution Infection period Diagnostic Cycle Species Group Denmark, Netherland, Incubation period : 3-4 month Ostrea edulis, O. Parasite of oyster haemocytes Throughout the year France, Ireland, Great Britain in infected area. Histology, Bonamia ostreae Protozoan Conchaphila, O. Angasi, O. (=>all the tissues can be invaded). (with a peak in Direct (except Scotland), Italy, tissue imprint, PCR, ISH, Puelchana, Tiostrea chilensis Oyster mortality September) Spain and USA electron microscopy Parasite of oyster haemocytes Throughout the year Histology, tissue imprint, Ostrea angasi, Ostrea Australia, New Zéland, Bonamia exitiosa Protozoan (=>all the tissues can be invaded). (with a peak during PCR, ISH, electron Unknown chilensis, Ostrea edulis Tasmania, Spain (in 2007) Oyster mortality Australian autumn) microscopy Parasites present in connective Histology, tissue imprint, Haplosporidium tissue (mantle, gonads, digestive Protozoan Crassostrea virginica USA, Canada Spring-summer PCR, ISH, electron Unknown costale gland) but not in digestive tubule microscopy epithelia. Mortality in May-June Parasites present in gills, connective tissue. Sporulation only Histology, tissue imprint, Unknown but Haplosporidium Crassostrea virginica, USA, Canada, Japan, Korea, Summer (May until Protozoan in the epithelium of digestive gland smear, PCR, HIS, electron intermediate host is nelsoni Crassostrea gigas France October) in C. virginica, Mortalities in C. microscopy suspected virginica. No mortality in C. gigas Parasite present in connective Haplosporidium Protozoan O. edulis, O. angasi tissue. Sometimes, mortality can France, Netherland, Spain Histology, tissue imprint Unknown armoricanum be observed Ostrea edulis , Tiostrea Spring-summer Histology, tissue imprint, ISH, Intermediate host: Extracellular parasite of the Marteilia refringens Protozoan chilensis, O. -

Breeding and Domestication of Eastern Oyster (Crassostrea
W&M ScholarWorks VIMS Articles Virginia Institute of Marine Science 2014 Breeding And Domestication Of Eastern Oyster (Crassostrea Virginica) Lines For Culture In The Mid-Atlantic, Usa: Line Development And Mass Selection For Disease Resistance Anu Frank-Lawale Virginia Institute of Marine Science Standish K. Allen Jr. Virginia Institute of Marine Science Lionel Degremont Virginia Institute of Marine Science Follow this and additional works at: https://scholarworks.wm.edu/vimsarticles Part of the Marine Biology Commons Recommended Citation Frank-Lawale, Anu; Allen, Standish K. Jr.; and Degremont, Lionel, "Breeding And Domestication Of Eastern Oyster (Crassostrea Virginica) Lines For Culture In The Mid-Atlantic, Usa: Line Development And Mass Selection For Disease Resistance" (2014). VIMS Articles. 334. https://scholarworks.wm.edu/vimsarticles/334 This Article is brought to you for free and open access by the Virginia Institute of Marine Science at W&M ScholarWorks. It has been accepted for inclusion in VIMS Articles by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. Journal of Shellfish Research, Vol. 33, No. 1, 153–165, 2014. BREEDING AND DOMESTICATION OF EASTERN OYSTER (CRASSOSTREA VIRGINICA) LINES FOR CULTURE IN THE MID-ATLANTIC, USA: LINE DEVELOPMENT AND MASS SELECTION FOR DISEASE RESISTANCE ANU FRANK-LAWALE,* STANDISH K. ALLEN, JR. AND LIONEL DE´GREMONT† Virginia Institute of Marine Science, Aquaculture Genetics and Breeding Technology Center, College of William and Mary, 1375 Greate Road, Gloucester Point, VA 23062 ABSTRACT A selective breeding program for Crassostrea virginica was established in 1997 as part of an initiative in Virginia to address declining oyster harvests caused by the two oyster pathogens Haplosporidium nelsoni (MSX) and Perkinsus marinus (Dermo). -

Structure of Protistan Parasites Found in Bivalve Molluscs
W&M ScholarWorks VIMS Books and Book Chapters Virginia Institute of Marine Science 1988 Structure of Protistan Parasites Found in Bivalve Molluscs Frank O. Perkins Follow this and additional works at: https://scholarworks.wm.edu/vimsbooks Part of the Marine Biology Commons, and the Parasitology Commons American Fisheries Society Special Publication 18:93- 111 , 1988 CC> Copyrighl by !he American Fisheries Sociely 1988 PARASITE MORPHOLOGY, STRATEGY, AND EVOLUTION Structure of Protistan Parasites Found in Bivalve Molluscs 1 FRANK 0. PERKINS Virginia In stitute of Marine Science. School of Marine Science, College of William and Mary Gloucester Point, Virginia 23062, USA Abstral'I.-The literature on the structure of protists parasitizing bivalve molluscs is reviewed, and previously unpubli shed observations of species of class Perkinsea, phylum Haplosporidia, and class Paramyxea are presented. Descriptions are given of the flagellar apparatus of Perkin.His marinus zoospores, the ultrastructure of Perkinsus sp. from the Baltic macoma Maconw balthica, and the development of haplosporosome-like bodies in Haplosporidium nelsoni. The possible origin of stem cells of Marreilia sydneyi from the inner two sporoplasms is discussed. New research efforts are suggested which could help elucidate the phylogenetic interrelationships and taxonomic positions of the various taxa and help in efforts to better understand life cycles of selected species. Studies of the structure of protistan parasites terization of the parasite species, to elucidation of found in bivalve moll uscs have been fruitful to the many parasite life cycles, and to knowledge of morphologist interested in comparative morphol- parasite metabolism. The latter, especially, is ogy, evolu tion, and taxonomy. -

Shellfish Reefs at Risk
SHELLFISH REEFS AT RISK A Global Analysis of Problems and Solutions Michael W. Beck, Robert D. Brumbaugh, Laura Airoldi, Alvar Carranza, Loren D. Coen, Christine Crawford, Omar Defeo, Graham J. Edgar, Boze Hancock, Matthew Kay, Hunter Lenihan, Mark W. Luckenbach, Caitlyn L. Toropova, Guofan Zhang CONTENTS Acknowledgments ........................................................................................................................ 1 Executive Summary .................................................................................................................... 2 Introduction .................................................................................................................................. 6 Methods .................................................................................................................................... 10 Results ........................................................................................................................................ 14 Condition of Oyster Reefs Globally Across Bays and Ecoregions ............ 14 Regional Summaries of the Condition of Shellfish Reefs ............................ 15 Overview of Threats and Causes of Decline ................................................................ 28 Recommendations for Conservation, Restoration and Management ................ 30 Conclusions ............................................................................................................................ 36 References ............................................................................................................................. -

Ecological Consequences of Pre-Contact Harvesting of Bay of Islands Fish and Shellfish, and Other Marine Taxa, Based on Midden Evidence
Journal of Pacific Archaeology – Vol. 7 · No. 2 · 2016 – article – Ecological Consequences of Pre-Contact Harvesting of Bay of Islands Fish and Shellfish, and other Marine Taxa, based on Midden Evidence John D. Booth1 ABSTRACT Midden contents – especially those that have associated dates – can provide compelling evidence concerning the effects of human harvesting on the diversity, distribution, abundance, and mean individual-size of shallow-water marine stocks. Archaeological Site Recording Scheme Site Record Forms for the 767 Bay of Islands middens as of August 2014 were summarised according to contents; these included 28 calibrated dates associated with 16 individual sites. The oldest site was first settled possibly as early as the 13th Century. By the time of European contact, the population of the Bay of Islands was possibly as great as 10,000 (over half the resident population today), yet it seems that the 500 years of harvesting pressure left no lasting legacy on Bay of Islands’ fish and shellfish resources – with the probable exception of the fishing-out of local populations of the Cook Strait limpet, and possibly the overfishing of hapuku in shallow waters. Marine mammal and seabird bones were only reported from Early and Early/Middle Period middens, consistent with the rapid extirpation and extinction of taxa after human arrival in the northeast of the North Island. Keywords: Bay of Islands, middens, fish, shellfish, seabird, marine mammal, ecological impact INTRODUCTION eri and Waikino inlets (Figure 1) – later mined and kiln- burnt to sweeten local soils (Nevin 1984; NAR 2004) – were Māori were prodigious consumers of fish and shellfish, so so prominent as to be singled out in the 1922 geological much so that missionary William Colenso was moved to chart (Ferrar & Cropp 1922). -

Eastern Oyster (Crassostrea Virginica)
Eastern Oyster (Crassostrea virginica) Imagine yourself on the streets of Manhattan, hungry but short of time and money. You see a pushcart, place your order and are served a quick lunch of…..oysters! That’s right, oysters. Throughout the 19th and early 20th Centuries, New York City was an oyster-eating town with oyster barges lining the waterfront and oysters served and sold on the streets. The abundance of these tasty bivalves was a welcome food source for the Dutch and English colonists and oysters, exported back to Europe, quickly became a source of economic wealth. So many oysters were sold that paths and extended shorelines were built in New York City on crushed shells. Oysters have been a prominent species in the New York/New Jersey Harbor Estuary since the end of the Ice Age. They have been documented as a food source in the Estuary for as long as 8,000 years, based on evidence from Native American midden (trash) piles. Later, many of the Harbor Estuary’s shoreline communities developed and thrived on the oyster trade until it collapsed in the mid-1920s, although minor oyster fisheries survived at the Harbor Estuary’s Jamaica Bay fringes where the East River meets Long Island Sound until the late1930s or later. In the 1880’s it was estimated that oysters covered about 350 square miles or 250,000 acres of the Harbor Estuary’s bottom. They were found in mid-to lower salinity areas including the tidal rivers in New Jersey’s Monmouth County, Raritan Bay, up the lower Raritan River, throughout the Arthur Kill, Newark Bay, the lower Rahway, Passaic, and Hackensack Rivers, the Kill Van Kull, up both sides of the Hudson River into Haverstraw Bay, around New York City in the Harlem and East Rivers and in many smaller tributaries and Jamaica Bay. -

Olympia Oyster (Ostrea Lurida)
COSEWIC Assessment and Status Report on the Olympia Oyster Ostrea lurida in Canada SPECIAL CONCERN 2011 COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: COSEWIC. 2011. COSEWIC assessment and status report on the Olympia Oyster Ostrea lurida in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. xi + 56 pp. (www.sararegistry.gc.ca/status/status_e.cfm). Previous report(s): COSEWIC. 2000. COSEWIC assessment and status report on the Olympia Oyster Ostrea conchaphila in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. vii + 30 pp. (www.sararegistry.gc.ca/status/status_e.cfm) Gillespie, G.E. 2000. COSEWIC status report on the Olympia Oyster Ostrea conchaphila in Canada in COSEWIC assessment and update status report on the Olympia Oyster Ostrea conchaphila in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. 1-30 pp. Production note: COSEWIC acknowledges Graham E. Gillespie for writing the provisional status report on the Olympia Oyster, Ostrea lurida, prepared under contract with Environment Canada and Fisheries and Oceans Canada. The contractor’s involvement with the writing of the status report ended with the acceptance of the provisional report. Any modifications to the status report during the subsequent preparation of the 6-month interim and 2-month interim status reports were overseen by Robert Forsyth and Dr. Gerald Mackie, COSEWIC Molluscs Specialist Subcommittee Co-Chair. For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment Canada Ottawa, ON K1A 0H3 Tel.: 819-953-3215 Fax: 819-994-3684 E-mail: COSEWIC/[email protected] http://www.cosewic.gc.ca Également disponible en français sous le titre Ếvaluation et Rapport de situation du COSEPAC sur l’huître plate du Pacifique (Ostrea lurida) au Canada. -

Shellfish Hatchery
EAST HAMPTON TOWN SHELLFISH HATCHERY The 2015 Crew, left to right: Kate, Pete, Carissa, Shelby, and Barley 2015 ANNUAL REPORT AND 2016 OPERATING PLAN Prepared by Kate Rossi-Snook Edited by Barley Dunne East Hampton Town Shellfish Hatchery The skiff loaded for seeding in Lake Montauk Annual Report of Operations Mission Statement With a hatchery on Fort Pond Bay, a nursery on Three Mile Harbor, and a floating raft field growout system in Napeague Harbor, the East Hampton Town Shellfish Hatchery produces large quantities of oyster (Crassostrea virginica), clam (Mercenaria mercenaria), and bay scallop (Argopecten irradians) seed to enhance valuable shellfish stocks in local waterways. Shellfish are available for harvest by all permitted town residents. Cooperative research and experimentation concerning shellfish culture, the subsequent success of seed in the wild, and the status of the resource is undertaken and reported upon regularly, often funded and validated by scientific research grants. Educational opportunities afforded by the work include school group and open house tours and educational displays at community functions. Annual reporting includes production statistics and values, seed dissemination information, results of research initiatives, a summary of outreach efforts, the status of current and developing infrastructure, and a plan for the following year’s operations. 2015 Full-time Staff Part-time and Contractual Volunteers John “Barley” Dunne – Director Carissa Maurin – Environmental Aide Romy Macari Kate Rossi-Snook – Hatchery Manager Shelby Joyce – Environmental Aide (summer) Christopher Fox-Strauss Pete Topping – Algae Culturist Adam Younes – Environmental Aide (fall) Jeremy Gould – Maintenance Mechanic Carissa and Pete unloading OysterGros Special Thanks to: Barnaby Friedman for producing our annual seeding maps. -

Background the Eastern Oyster, Crassostrea Virginica, Is An
Background The eastern oyster, Crassostrea virginica, is an important keystone species in Great Bay Estuary, NH. As an ecosystem engineer, oysters provide several ecosystem services to both people and wildlife. Oysters filter excess nutrients and suspended solids from the water column improving water quality and clarity (Coen et al., 2007). In addition, oyster reefs provide important habitat for fish and invertebrates by building large vertical complex reef structures (Coen et al., 2007). Historically, Great Bay Estuary was filled with acres of healthy oyster reef. However, due to pollution, disease, sedimentation, and historical harvesting these numbers have decreased by over 90% resulting in only a little over a 100 acres of oyster reef today. With this drastic loss of oyster reefs, Great Bay has experienced a similar loss in the important ecosystem services that oysters provide to estuarine ecosystems. For this reason, The Nature Conservancy (TNC) of New Hampshire has been working collaboratively with The University of New Hampshire’s Jackson Estuarine Laboratory (UNH-JEL) to restore oyster reefs to Great Bay since 2009. The Oyster Conservationist (OC) Program is an important community engagement component of oyster reef restoration in Great Bay. An Oyster Conservationist is a community member and environmental steward in the coastal area of New Hampshire who advocates or acts for the protection and preservation of the environment and wildlife. Participants in the OC Program work towards improving the health of Great Bay by raising oyster spat for TNC’s oyster reef restoration projects. Volunteers adopt a cage with spat on shell for an eight-week period cleaning and caring for the cage while also collecting data throughout the summer on survival, growth, invasive species, and wild oyster spat settlement. -

The Eastern Oyster: a Not-So-Typical Mollusc Lab Investigation: Class Bivalvia High School Version Lesson by Kevin Goff
The Eastern Oyster: A Not-So-Typical Mollusc Lab Investigation: Class Bivalvia High School Version Lesson by Kevin Goff SETTING THE STAGE The earliest animals on Earth had either irregular, asymmetrical VIDEOS TO WATCH bodies or radial symmetry, with a body shaped like a merry-go- For dazzling displays of how asymmetri- round. Animals with these body plans usually sit still on the seafloor cal and radial animals can harvest food – like sponges, coral, and sea anemone. Others – like jellyfish – just from any direction, visit the Shape of Life drift along on ocean currents. These animals do not actively forage website and watch these two clips (under for food. Instead, they wait for food to come to them. Their body “Behavior”): • Sponges: Filter Feeding Made Visible shape lets them collect food from any direction. Eventually, though, (2.5 min) a line of worm-like animals evolved bilateral symmetry, with a body • Cnidarians: Anemone Catches Goby bearing two sides – left versus right – that are mirror images of each (2.5 min) other. This body plan is an adaptation for directional movement. To understand why, just imagine a car with monster truck tires on one side and little red wagon wheels on the other. It would go in circles! Having identical left and right halves enables an animal to track in a straight line. Animals with bilateral symmetry also usually have a distinct head VIDEOS TO WATCH at one end, where the mouth and sense organs are concentrated. To see how a bilateral, cephalized body We say they are cephalized, meaning “head-having.” In contrast, lets an animal actively seek food, animals with radial symmetry are not cephalized: They have watch these two Shape of Life clips: • Flatworm Animation: Body Plan no head, just a mouth in the middle.