Discretization for Naive-Bayes Learning: Managing Discretization Bias and Variance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

On the Use of Discrete Fourier Transform for Solving Biperiodic Boundary Value Problem of Biharmonic Equation in the Unit Rectangle
On the Use of Discrete Fourier Transform for Solving Biperiodic Boundary Value Problem of Biharmonic Equation in the Unit Rectangle Agah D. Garnadi 31 December 2017 Department of Mathematics, Faculty of Mathematics and Natural Sciences, Bogor Agricultural University Jl. Meranti, Kampus IPB Darmaga, Bogor, 16680 Indonesia Abstract This note is addressed to solving biperiodic boundary value prob- lem of biharmonic equation in the unit rectangle. First, we describe the necessary tools, which is discrete Fourier transform for one di- mensional periodic sequence, and then extended the results to 2- dimensional biperiodic sequence. Next, we use the discrete Fourier transform 2-dimensional biperiodic sequence to solve discretization of the biperiodic boundary value problem of Biharmonic Equation. MSC: 15A09; 35K35; 41A15; 41A29; 94A12. Key words: Biharmonic equation, biperiodic boundary value problem, discrete Fourier transform, Partial differential equations, Interpola- tion. 1 Introduction This note is an extension of a work by Henrici [2] on fast solver for Poisson equation to biperiodic boundary value problem of biharmonic equation. This 1 note is organized as follows, in the first part we address the tools needed to solve the problem, i.e. introducing discrete Fourier transform (DFT) follows Henrici's work [2]. The second part, we apply the tools already described in the first part to solve discretization of biperiodic boundary value problem of biharmonic equation in the unit rectangle. The need to solve discretization of biharmonic equation is motivated by its occurence in some applications such as elasticity and fluid flows [1, 4], and recently in image inpainting [3]. Note that the work of Henrici [2] on fast solver for biperiodic Poisson equation can be seen as solving harmonic inpainting. -
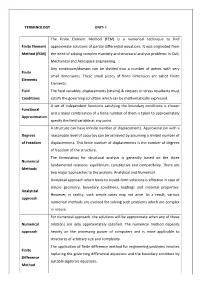
TERMINOLOGY UNIT- I Finite Element Method (FEM)
TERMINOLOGY UNIT- I The Finite Element Method (FEM) is a numerical technique to find Finite Element approximate solutions of partial differential equations. It was originated from Method (FEM) the need of solving complex elasticity and structural analysis problems in Civil, Mechanical and Aerospace engineering. Any continuum/domain can be divided into a number of pieces with very Finite small dimensions. These small pieces of finite dimension are called Finite Elements Elements. Field The field variables, displacements (strains) & stresses or stress resultants must Conditions satisfy the governing condition which can be mathematically expressed. A set of independent functions satisfying the boundary conditions is chosen Functional and a linear combination of a finite number of them is taken to approximately Approximation specify the field variable at any point. A structure can have infinite number of displacements. Approximation with a Degrees reasonable level of accuracy can be achieved by assuming a limited number of of Freedom displacements. This finite number of displacements is the number of degrees of freedom of the structure. The formulation for structural analysis is generally based on the three Numerical fundamental relations: equilibrium, constitutive and compatibility. There are Methods two major approaches to the analysis: Analytical and Numerical. Analytical approach which leads to closed-form solutions is effective in case of simple geometry, boundary conditions, loadings and material properties. Analytical However, in reality, such simple cases may not arise. As a result, various approach numerical methods are evolved for solving such problems which are complex in nature. For numerical approach, the solutions will be approximate when any of these Numerical relations are only approximately satisfied. -

Lecture 7: the Complex Fourier Transform and the Discrete Fourier Transform (DFT)
Lecture 7: The Complex Fourier Transform and the Discrete Fourier Transform (DFT) c Christopher S. Bretherton Winter 2014 7.1 Fourier analysis and filtering Many data analysis problems involve characterizing data sampled on a regular grid of points, e. g. a time series sampled at some rate, a 2D image made of regularly spaced pixels, or a 3D velocity field from a numerical simulation of fluid turbulence on a regular grid. Often, such problems involve characterizing, detecting, separating or ma- nipulating variability on different scales, e. g. finding a weak systematic signal amidst noise in a time series, edge sharpening in an image, or quantifying the energy of motion across the different sizes of turbulent eddies. Fourier analysis using the Discrete Fourier Transform (DFT) is a fun- damental tool for such problems. It transforms the gridded data into a linear combination of oscillations of different wavelengths. This partitions it into scales which can be separately analyzed and manipulated. The computational utility of Fourier methods rests on the Fast Fourier Transform (FFT) algorithm, developed in the 1960s by Cooley and Tukey, which allows efficient calculation of discrete Fourier coefficients of a periodic function sampled on a regular grid of 2p points (or 2p3q5r with slightly reduced efficiency). 7.2 Example of the FFT Using Matlab, take the FFT of the HW1 wave height time series (length 24 × 60=25325) and plot the result (Fig. 1): load hw1 dat; zhat = fft(z); plot(abs(zhat),’x’) A few elements (the second and last, and to a lesser extent the third and the second from last) have magnitudes that stand above the noise. -

Discretization of Integro-Differential Equations
Discretization of Integro-Differential Equations Modeling Dynamic Fractional Order Viscoelasticity K. Adolfsson1, M. Enelund1,S.Larsson2,andM.Racheva3 1 Dept. of Appl. Mech., Chalmers Univ. of Technology, SE–412 96 G¨oteborg, Sweden 2 Dept. of Mathematics, Chalmers Univ. of Technology, SE–412 96 G¨oteborg, Sweden 3 Dept. of Mathematics, Technical University of Gabrovo, 5300 Gabrovo, Bulgaria Abstract. We study a dynamic model for viscoelastic materials based on a constitutive equation of fractional order. This results in an integro- differential equation with a weakly singular convolution kernel. We dis- cretize in the spatial variable by a standard Galerkin finite element method. We prove stability and regularity estimates which show how the convolution term introduces dissipation into the equation of motion. These are then used to prove a priori error estimates. A numerical ex- periment is included. 1 Introduction Fractional order operators (integrals and derivatives) have proved to be very suit- able for modeling memory effects of various materials and systems of technical interest. In particular, they are very useful when modeling viscoelastic materials, see, e.g., [3]. Numerical methods for quasistatic viscoelasticity problems have been studied, e.g., in [2] and [8]. The drawback of the fractional order viscoelastic models is that the whole strain history must be saved and included in each time step. The most commonly used algorithms for this integration are based on the Lubich convolution quadrature [5] for fractional order operators. In [1, 2], we develop an efficient numerical algorithm based on sparse numerical quadrature earlier studied in [6]. While our earlier work focused on discretization in time for the quasistatic case, we now study space discretization for the fully dynamic equations of mo- tion, which take the form of an integro-differential equation with a weakly singu- lar convolution kernel. -

There Is Only One Fourier Transform Jens V
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 25 December 2017 doi:10.20944/preprints201712.0173.v1 There is only one Fourier Transform Jens V. Fischer * Microwaves and Radar Institute, German Aerospace Center (DLR), Germany Abstract Four Fourier transforms are usually defined, the Integral Fourier transform, the Discrete-Time Fourier transform (DTFT), the Discrete Fourier transform (DFT) and the Integral Fourier transform for periodic functions. However, starting from their definitions, we show that all four Fourier transforms can be reduced to actually only one Fourier transform, the Fourier transform in the distributional sense. Keywords Integral Fourier Transform, Discrete-Time Fourier Transform (DTFT), Discrete Fourier Transform (DFT), Integral Fourier Transform for periodic functions, Fourier series, Poisson Summation Formula, periodization trick, interpolation trick Introduction Two Important Operations The fact that “there is only one Fourier transform” actually There are two operations which are being strongly related to needs no proof. It is commonly known and often discussed in four different variants of the Fourier transform, i.e., the literature, for example in [1-2]. discretization and periodization. For any real-valued T > 0 and δ(t) being the Dirac impulse, let However frequently asked questions are: (1) What is the difference between calculating the Fourier series and the (1) Fourier transform of a continuous function? (2) What is the difference between Discrete-Time Fourier Transform (DTFT) be the function that results from a discretization of f(t) and let and Discrete Fourier Transform (DFT)? (3) When do we need which Fourier transform? and many others. The default answer (2) today to all these questions is to invite the reader to have a look at the so-called Fourier-Poisson cube, e.g. -

Data Mining Algorithms
Data Management and Exploration © for the original version: Prof. Dr. Thomas Seidl Jiawei Han and Micheline Kamber http://www.cs.sfu.ca/~han/dmbook Data Mining Algorithms Lecture Course with Tutorials Wintersemester 2003/04 Chapter 4: Data Preprocessing Chapter 3: Data Preprocessing Why preprocess the data? Data cleaning Data integration and transformation Data reduction Discretization and concept hierarchy generation Summary WS 2003/04 Data Mining Algorithms 4 – 2 Why Data Preprocessing? Data in the real world is dirty incomplete: lacking attribute values, lacking certain attributes of interest, or containing only aggregate data noisy: containing errors or outliers inconsistent: containing discrepancies in codes or names No quality data, no quality mining results! Quality decisions must be based on quality data Data warehouse needs consistent integration of quality data WS 2003/04 Data Mining Algorithms 4 – 3 Multi-Dimensional Measure of Data Quality A well-accepted multidimensional view: Accuracy (range of tolerance) Completeness (fraction of missing values) Consistency (plausibility, presence of contradictions) Timeliness (data is available in time; data is up-to-date) Believability (user’s trust in the data; reliability) Value added (data brings some benefit) Interpretability (there is some explanation for the data) Accessibility (data is actually available) Broad categories: intrinsic, contextual, representational, and accessibility. WS 2003/04 Data Mining Algorithms 4 – 4 Major Tasks in Data Preprocessing -

Verification and Validation in Computational Fluid Dynamics1
SAND2002 - 0529 Unlimited Release Printed March 2002 Verification and Validation in Computational Fluid Dynamics1 William L. Oberkampf Validation and Uncertainty Estimation Department Timothy G. Trucano Optimization and Uncertainty Estimation Department Sandia National Laboratories P. O. Box 5800 Albuquerque, New Mexico 87185 Abstract Verification and validation (V&V) are the primary means to assess accuracy and reliability in computational simulations. This paper presents an extensive review of the literature in V&V in computational fluid dynamics (CFD), discusses methods and procedures for assessing V&V, and develops a number of extensions to existing ideas. The review of the development of V&V terminology and methodology points out the contributions from members of the operations research, statistics, and CFD communities. Fundamental issues in V&V are addressed, such as code verification versus solution verification, model validation versus solution validation, the distinction between error and uncertainty, conceptual sources of error and uncertainty, and the relationship between validation and prediction. The fundamental strategy of verification is the identification and quantification of errors in the computational model and its solution. In verification activities, the accuracy of a computational solution is primarily measured relative to two types of highly accurate solutions: analytical solutions and highly accurate numerical solutions. Methods for determining the accuracy of numerical solutions are presented and the importance of software testing during verification activities is emphasized. The fundamental strategy of 1Accepted for publication in the review journal Progress in Aerospace Sciences. 3 validation is to assess how accurately the computational results compare with the experimental data, with quantified error and uncertainty estimates for both. -

High Order Regularization of Dirac-Delta Sources in Two Space Dimensions
INTERNSHIP REPORT High order regularization of Dirac-delta sources in two space dimensions. Author: Supervisors: Wouter DE VRIES Prof. Guustaaf JACOBS s1010239 Jean-Piero SUAREZ HOST INSTITUTION: San Diego State University, San Diego (CA), USA Department of Aerospace Engineering and Engineering Mechanics Computational Fluid Dynamics laboratory Prof. G.B. JACOBS HOME INSTITUTION University of Twente, Enschede, Netherlands Faculty of Engineering Technology Group of Engineering Fluid Dynamics Prof. H.W.M. HOEIJMAKERS March 4, 2015 Abstract In this report the regularization of singular sources appearing in hyperbolic conservation laws is studied. Dirac-delta sources represent small particles which appear as source-terms in for example Particle-laden flow. The Dirac-delta sources are regularized with a high order accurate polynomial based regularization technique. Instead of regularizing a single isolated particle, the aim is to employ the regularization technique for a cloud of particles distributed on a non- uniform grid. By assuming that the resulting force exerted by a cloud of particles can be represented by a continuous function, the influence of the source term can be approximated by the convolution of the continuous function and the regularized Dirac-delta function. It is shown that in one dimension the distribution of the singular sources, represented by the convolution of the contin- uous function and Dirac-delta function, can be approximated with high accuracy using the regularization technique. The method is extended to two dimensions and high order approximations of the source-term are obtained as well. The resulting approximation of the singular sources are interpolated using a polynomial interpolation in order to regain a continuous distribution representing the force exerted by the cloud of particles. -

On Convergence of the Immersed Boundary Method for Elliptic Interface Problems
On convergence of the immersed boundary method for elliptic interface problems Zhilin Li∗ January 26, 2012 Abstract Peskin's Immersed Boundary (IB) method is one of the most popular numerical methods for many years and has been applied to problems in mathematical biology, fluid mechanics, material sciences, and many other areas. Peskin's IB method is associated with discrete delta functions. It is believed that the IB method is first order accurate in the L1 norm. But almost no rigorous proof could be found in the literature until recently [14] in which the author showed that the velocity is indeed first order accurate for the Stokes equations with a periodic boundary condition. In this paper, we show first order convergence with a log h factor of the IB method for elliptic interface problems essentially without the boundary condition restrictions. The results should be applicable to the IB method for many different situations involving elliptic solvers for Stokes and Navier-Stokes equations. keywords: Immersed Boundary (IB) method, Dirac delta function, convergence of IB method, discrete Green function, discrete Green's formula. AMS Subject Classification 2000 65M12, 65M20. 1 Introduction Since its invention in 1970's, the Immersed Boundary (IB) method [15] has been applied almost everywhere in mathematics, engineering, biology, fluid mechanics, and many many more areas, see for example, [16] for a review and references therein. The IB method is not only a mathematical modeling tool in which a complicated boundary condition can be treated as a source distribution but also a numerical method in which a discrete delta function is used. -

Domain Decomposition and Discretization of Continuous Mathematical Models
An International Journal computers & mathematics with applications PERGAMON Computers and Mathematics with Applications 38 (1999) 79-88 www.elsevier.nl/locate/camwa Domain Decomposition and Discretization of Continuous Mathematical Models I. BONZANI Department of Mathematics, Potitecnico, Torino, Italy Bonzani©Polito. it (Received April 1999; accepted May 1999) Abstract--This paper deals with the development of decomposition of domains methods related to the discretization, by collocation-interpolation methods, of continuous models described by nonlinear partial differential equations. The objective of this paper is to show how generalized collocation and domain decomposition methods can be applied for problems where different models are used within the same domain. © 1999 Elsevier Science Ltd. All rights reserved. Keywords--Nonlinear sciences, Evolution equations, Sinc functions, Domain decomposition, Dif- ferential quadrature. 1. INTRODUCTION A standard solution technique of nonlinear initial-boundary value problem for partial differential equations is the generalized collocation-interpolation method, originally proposed as differential quadrature method. This method, as documented in [1], as well as in some more recent devel- opments [2,3], discretizes the original continuous model (and problem) into a discrete (in space) model, with a finite number of degrees of freedom, while the initial-boundary value problem is transformed into an initial-value problem for ordinary differential equations. This method is well documented in the literature on applied mathematics. It was proposed by Bellman and Casti [1] and developed by several authors in the deterministic and stochastic framework as documented in Chapters 3 and 4 of [4]. The validity of the method, with respect to alternative ones, e.g., finite elements and volumes [5,6], Trefftz methods [7,8], spectral meth- ods [9,10], is not here discussed. -

Detailed Comparison of Numerical Methods for the Perturbed Sine-Gordon Equation with Impulsive Forcing
J Eng Math (2014) 87:167–186 DOI 10.1007/s10665-013-9678-x Detailed comparison of numerical methods for the perturbed sine-Gordon equation with impulsive forcing Danhua Wang · Jae-Hun Jung · Gino Biondini Received: 5 September 2012 / Accepted: 25 October 2013 / Published online: 20 March 2014 © Springer Science+Business Media Dordrecht 2014 Abstract The properties of various numerical methods for the study of the perturbed sine-Gordon (sG) equation with impulsive forcing are investigated. In particular, finite difference and pseudo-spectral methods for discretizing the equation are considered. Different methods of discretizing the Dirac delta are discussed. Various combinations of these methods are then used to model the soliton–defect interaction. A comprehensive study of convergence of all these combinations is presented. Detailed explanations are provided of various numerical issues that should be carefully considered when the sG equation with impulsive forcing is solved numerically. The properties of each method depend heavily on the specific representation chosen for the Dirac delta—and vice versa. Useful comparisons are provided that can be used for the design of the numerical scheme to study the singularly perturbed sG equation. Some interesting results are found. For example, the Gaussian approximation yields the worst results, while the domain decomposition method yields the best results, for both finite difference and spectral methods. These findings are corroborated by extensive numerical simulations. Keywords Finite difference methods · Impulsive forcing · Sine-Gordon equation · Spectral methods 1 Introduction The sine-Gordon (sG) equation is a ubiquitous physical model that describes nonlinear oscillations in various settings such as Josephson junctions, self-induced transparency, crystal dislocations, Bloch wall motion of magnetic crystals, and beyond (see [1] for references). -

Peridigm Users Guide
Peridigm DLR-IB-FA-BS-2018-23 Peridigm Users Guide For Peridigm versions 1.4.1 Martin Rädel DLR German Aerospace Center Composite Structures and Adaptive Systems Structural Mechanics Braunschweig Deutsches Zentrum DLR für Luft- und Raumfahrt German Aerospace Center Document Identification ii DLR German Aerospace Center Composite Structures and Adaptive Systems Structural Mechanics Dr. Tobias Wille Lilienthalplatz 7 38108 Braunschweig Germany Tel: +49 531 295-3012 Fax: +49 531 295-2232 Web: http://www.dlr.de/fa/en Martin Rädel Tel: +49 531 295-2048 Fax: +49 531 295-2232 Mail: [email protected] Document Identification: Report number . DLR-IB-FA-BS-2018-23 Title . Peridigm Users Guide Subject . Peridigm Author(s) . Martin Rädel With contributions by . Christian Willberg Filename . Peridigm_Users_Guide.tex Last saved by . DLR\raed_ma Last saved on . 30th October 2018 Document History: Version 0.0.1 . Initial draft 19.02.2016 Version 0.0.2 . A lot of additions 23.02.2017 Version 0.0.3 . Glossary, user output & results 17.05.2017 Version 0.0.4 . Index 27.09.2017 Version 1.0.0 . Initial release version 31.01.2018 DLR – DLR-IB-FA-BS-2018-23 DLR Copyright © 2018 German Aerospace Center (DLR) Permission is granted to copy, distribute and/or modify this document under the terms of the BSD Documentation License. A copy of the license is included in the section entitled “BSD Documentation License”. Dieses Dokument darf unter den Bedingungen der BSD Documentation License vervielfältigt, distribuiert und/oder modifiziert werden. Eine Kopie der Lizenz ist im Kapitel “BSD Docu- mentation License” enthalten.