From Neurospora Crassa
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

Eif3j Antibody A
Revision 1 C 0 2 - t eIF3J Antibody a e r o t S Orders: 877-616-CELL (2355) [email protected] Support: 877-678-TECH (8324) 1 6 Web: [email protected] 2 www.cellsignal.com 3 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source: UniProt ID: Entrez-Gene Id: WB, IP H M R Endogenous 35 Rabbit O75822 8669 Product Usage Information (5). eIF3J has been shown to associate with the aminoacyl site and mRNA entry channel of the 40S ribosomal subunit (6). eIF3J has also been shown to have an additional role in Application Dilution the recycling of post-termination complexes (7). 1. Masutani, M. et al. (2007) EMBO J 26, 3373-83. Western Blotting 1:1000 2. Chaudhuri, J. et al. (1999) J Biol Chem 274, 17975-80. Immunoprecipitation 1:50 3. Holz, M.K. et al. (2005) Cell 123, 569-80. 4. Harris, T.E. et al. (2006) EMBO J 25, 1659-68. 5. Fraser, C.S. et al. (2004) J Biol Chem 279, 8946-56. Storage 6. Fraser, C.S. et al. (2007) Mol Cell 26, 811-9. Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA and 50% 7. Pisarev, A.V. et al. (2007) Cell 131, 286-99. glycerol. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity eIF3J Antibody detects endogenous levels of total eIF3J protein. Species Reactivity: Human, Mouse, Rat Source / Purification Polyclonal antibodies are produced by immunizing animals with a synthetic peptide corresponding to the sequence of human eIF3J. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Gene Expression Profiling Analysis Contributes to Understanding the Association Between Non-Syndromic Cleft Lip and Palate, and Cancer
2110 MOLECULAR MEDICINE REPORTS 13: 2110-2116, 2016 Gene expression profiling analysis contributes to understanding the association between non-syndromic cleft lip and palate, and cancer HONGYI WANG, TAO QIU, JIE SHI, JIULONG LIANG, YANG WANG, LIANGLIANG QUAN, YU ZHANG, QIAN ZHANG and KAI TAO Department of Plastic Surgery, General Hospital of Shenyang Military Area Command, PLA, Shenyang, Liaoning 110016, P.R. China Received March 10, 2015; Accepted December 18, 2015 DOI: 10.3892/mmr.2016.4802 Abstract. The present study aimed to investigate the for NSCL/P were implicated predominantly in the TGF-β molecular mechanisms underlying non-syndromic cleft lip, signaling pathway, the cell cycle and in viral carcinogenesis. with or without cleft palate (NSCL/P), and the association The TP53, CDK1, SMAD3, PIK3R1 and CASP3 genes were between this disease and cancer. The GSE42589 data set found to be associated, not only with NSCL/P, but also with was downloaded from the Gene Expression Omnibus data- cancer. These results may contribute to a better understanding base, and contained seven dental pulp stem cell samples of the molecular mechanisms of NSCL/P. from children with NSCL/P in the exfoliation period, and six controls. Differentially expressed genes (DEGs) were Introduction screened using the RankProd method, and their potential functions were revealed by pathway enrichment analysis and Non-syndromic cleft lip, with or without cleft palate (NSCL/P) construction of a pathway interaction network. Subsequently, is one of the most common types of congenital defect and cancer genes were obtained from six cancer databases, and affects 3.4-22.9/10,000 individuals worldwide (1). -

Translation Initiation Factor Eif3h Targets Specific Transcripts To
Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis Avik Choudhuria,b, Umadas Maitraa,1, and Todd Evansb,1 aDepartment of Developmental and Molecular Biology, Albert Einstein College of Medicine of Yeshiva University, Bronx, NY 10461; and bDepartment of Surgery, Weill Cornell Medical College, New York, NY 10065 Edited by Igor B. Dawid, The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, and approved April 30, 2013 (received for review February 14, 2013) Eukaryotic translation initiation factor 3 (eIF3) plays a central role eukaryotes. These—eIF3d, eIF3e, eIF3f, eIF3h, eIF3j, eIF3k, in translation initiation and consists of five core (conserved) sub- eIF3l, and eIF3m—were designated “non-core” subunits (4). units present in both budding yeast and higher eukaryotes. Higher In contrast to the budding yeast, the genome of the fission eukaryotic eIF3 contains additional (noncore or nonconserved) yeast Schizosaccharomyces pombe contains structural homologs subunits of poorly defined function, including sub-unit h (eIF3h), of at least five noncore (nonconserved) eIF3 subunits—eIF3d, which in zebrafish is encoded by two distinct genes (eif3ha and eIF3e, eIF3f, eIF3h, and eIF3m. The gene encoding eIF3f is eif3hb). Previously we showed that eif3ha encodes the predominant essential for growth, whereas eIF3d, eIF3e, and eIF3h are dis- isoform during zebrafish embryogenesis and that depletion of this pensable for growth and viability (5–11). However, deleted factor causes defects in the development of the brain and eyes. To strains show specific phenotypes including defects in meiosis/ investigate the molecular mechanism governing this regulation, we sporulation (6, 9, 11). -

Supplemental Table 1. 3 Types of Signals That Are Involved in T Cell Activation
Supplemental Table 1. 3 types of signals that are involved in T cell activation Syn-T cells Allo-T cells CD3/28- T cells First signal Not engaged MHC-alloantigen- CD3-Engaged MHC-TCR-CD3 TCR-CD3 Engaged pathway Second signal Co- Not sure CD28, CTLA4, CD28 signaling stimulatory ICOS, OX40, CD30, molecules CD153 Third signal Not sure Cytokines from Allo- Cytokines from cytokines DCs and Allo-T cells CD3/28-T cells Supplementary Figure 1 Pearson Correlation Coefficient R A Exp.2 Exp.2 Exp.2 Syn Allo CD3/28 Exp.1 Exp.3 Exp.1 Exp.3 Exp.1 Exp.3 R=0.800981 R=0.815741 R=0.818577 Allo Allo Allo B Exp1 Exp2 Exp3 E Exp2 Syn CD3/28 Syn CD3/28 Syn CD3/28 R=0.6701227 R=0.6504327 R=0.6680361 Pearson Correlation Coefficient R for 9 biological replicates for affymatrix microarrays Allo C D Allo miRNA Exp1 Exp2 E Syn CD3/28 Syn CD3/28 R=0.938515(CD3/28) R=0.703578681 R=0.753278283 Pearson Correlation Coefficient R for 6 biological replicates for miRNA microarrays Supplementary Table 2 .Enrichments of Transcripts differentially Increased in Allo- T cells Fold Ch. Fold Ch. Fold Ch. Fold Fold Symbol ( Log) Symbol ( Log) Symbol (Log) Symbol Cha.(log) Symbol Ch.(Log) Alg8 2.060308 Cpsf6 1.22965 Arl6ip6 1.145309 5730471H19Rik 1.081846 Gmfb 1.029386 Melk 1.980582 Pno1 1.22959 C80350 1.143319 Grsf1 1.081728 Psmb7 1.028187 Eif3j 1.939782 BF228097 1.22056 Eif4a1 1.13752 Lmnb1 1.073975 Gltp 1.027779 Rg9mtd2 1.81345 Oxnad1 1.20962 Appbp2 1.137063 Senp1 1.073847 Fundc2 1.0272 Plk4 1.791148 BF228097 1.20522 Prkaca 1.136359 Psmd6 1.072738 Narfl 1.026824 Ikzf4 1.759402 -

Relevance of Translation Initiation in Diffuse Glioma Biology and Its
cells Review Relevance of Translation Initiation in Diffuse Glioma Biology and its Therapeutic Potential Digregorio Marina 1, Lombard Arnaud 1,2, Lumapat Paul Noel 1, Scholtes Felix 1,2, Rogister Bernard 1,3 and Coppieters Natacha 1,* 1 Laboratory of Nervous System Disorders and Therapy, GIGA-Neurosciences Research Centre, University of Liège, 4000 Liège, Belgium; [email protected] (D.M.); [email protected] (L.A.); [email protected] (L.P.N.); [email protected] (S.F.); [email protected] (R.B.) 2 Department of Neurosurgery, CHU of Liège, 4000 Liège, Belgium 3 Department of Neurology, CHU of Liège, 4000 Liège, Belgium * Correspondence: [email protected] Received: 18 October 2019; Accepted: 26 November 2019; Published: 29 November 2019 Abstract: Cancer cells are continually exposed to environmental stressors forcing them to adapt their protein production to survive. The translational machinery can be recruited by malignant cells to synthesize proteins required to promote their survival, even in times of high physiological and pathological stress. This phenomenon has been described in several cancers including in gliomas. Abnormal regulation of translation has encouraged the development of new therapeutics targeting the protein synthesis pathway. This approach could be meaningful for glioma given the fact that the median survival following diagnosis of the highest grade of glioma remains short despite current therapy. The identification of new targets for the development of novel therapeutics is therefore needed in order to improve this devastating overall survival rate. This review discusses current literature on translation in gliomas with a focus on the initiation step covering both the cap-dependent and cap-independent modes of initiation. -

Nat1 Promotes Translation of Specific Proteins That Induce Differentiation of Mouse Embryonic Stem Cells
Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells Hayami Sugiyamaa, Kazutoshi Takahashia,b, Takuya Yamamotoa,c,d, Mio Iwasakia, Megumi Naritaa, Masahiro Nakamuraa, Tim A. Randb, Masato Nakagawaa, Akira Watanabea,c,e, and Shinya Yamanakaa,b,1 aDepartment of Life Science Frontiers, Center for iPS Cell Research and Application, Kyoto University, Kyoto 606-8507, Japan; bGladstone Institute of Cardiovascular Disease, San Francisco, CA 94158; cInstitute for Integrated Cell-Material Sciences, Kyoto University, Kyoto 606-8501, Japan; dJapan Agency for Medical Research and Development-Core Research for Evolutional Science and Technology (AMED-CREST), Tokyo 100-0004, Japan; and eJapan Science and Technology Agency (JST)-CREST, Saitama 332-0012, Japan Contributed by Shinya Yamanaka, November 22, 2016 (sent for review October 18, 2016; reviewed by Katsura Asano and Keisuke Kaji) Novel APOBEC1 target 1 (Nat1) (also known as “p97,”“Dap5,” and To study the role of Nat1 in cell differentiation further, we “Eif4g2”) is a ubiquitously expressed cytoplasmic protein that is homol- generated mouse embryonic stem cells (mES cells) lacking both ogous to the C-terminal two thirds of eukaryotic translation initiation alleles of the Nat1 gene. mES cells were derived from blastocysts factor 4G (Eif4g1). We previously showed that Nat1-null mouse embry- in 1981 (11, 12) and possess two unique properties. First, ES cells onic stem cells (mES cells) are resistant to differentiation. In the current have the potential to self-renew indefinitely (maintenance). Sec- study, we found that NAT1 and eIF4G1 share many binding proteins, ond, ES cells have the potential to differentiate into all somatic such as the eukaryotic translation initiation factors eIF3 and eIF4A and and germ cell types (pluripotency) that make up the body. -

Release of Initiation Factors from 48S Complexes
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP Anett Unbehaun,1 Sergei I. Borukhov,1 Christopher U.T. Hellen,1 and Tatyana V. Pestova1,2,3 1Department of Microbiology, State University of New York Downstate Medical Center, Brooklyn, New York 11203, USA; 2A.N. Belozersky Institute of Physico-chemical Biology, Moscow State University, Moscow 119899, Russia. The 40S subunit in 48S complexes formed at the initiation codon of mRNA is bound to eukaryotic initiation Met factor (eIF) 3, eIF1, eIF1A, and an eIF2/GTP/Met-tRNAi ternary complex and can therefore not join a 60S subunit directly to form an 80S ribosome. We report that eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes led to release of eIF2-GDP but not eIF3 or eIF1. eIF5B did not influence factor release in the absence of 60S subunits. Therefore eIF3 and eIF1 dissociate from 40S subunits during, rather than before, the eIF5B-mediated subunit joining event. In the absence of eIF1, eIF5-stimulated hydrolysis of eIF2-bound GTP occurred at the same rate in 43S pre-initiation and 48S initiation complexes. GTP hydrolysis in 43S complexes assembled with eIF1 was much slower than in 43S or 48S complexes assembled without eIF1. Establishment of codon–anticodon base-pairing in 48S complexes relieved eIF1’s inhibition. Thus, in addition to its role in initiation codon selection during 48S complex formation, eIF1 also participates in maintaining the fidelity of the initiation process at a later stage, hydrolysis of eIF2-bound GTP, by inhibiting premature GTP hydrolysis and by linking establishment of codon–anticodon base-pairing with GTP hydrolysis. -
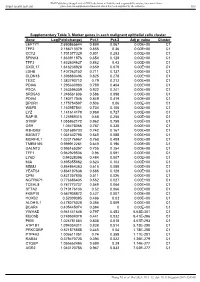
Value Cluster LEFTY1 2.690865644 0.899 0.067 0.00E+00 C1
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Supplementary Table 3. Marker genes in each malignant epithelial cells cluster Gene Log(Fold-change) Pct.1 Pct.2 Adj p- value Cluster LEFTY1 2.690865644 0.899 0.067 0.00E+00 C1 TFF2 2.186713379 0.855 0.36 0.00E+00 C1 CCT2 1.701377329 0.801 0.283 0.00E+00 C1 SPINK4 1.663911876 0.654 0.128 0.00E+00 C1 TFF3 1.632604627 0.882 0.43 0.00E+00 C1 CXCL17 1.616238928 0.659 0.079 0.00E+00 C1 LDHB 1.407263152 0.711 0.137 0.00E+00 C1 CLDN18 1.398880496 0.825 0.278 0.00E+00 C1 TESC 1.382790712 0.79 0.212 0.00E+00 C1 PDIA6 1.295630983 0.789 0.404 0.00E+00 C1 PSCA 1.263596359 0.922 0.241 0.00E+00 C1 SRD5A3 1.246561606 0.586 0.098 0.00E+00 C1 PDIA4 1.180717046 0.849 0.419 0.00E+00 C1 DPCR1 1.175754587 0.506 0.06 0.00E+00 C1 WBP5 1.163957541 0.734 0.106 0.00E+00 C1 LYZ 1.141614179 0.969 0.727 0.00E+00 C1 RAP1B 1.125989315 0.68 0.255 0.00E+00 C1 S100P 1.055862172 0.962 0.758 0.00E+00 C1 OS9 1.05478066 0.787 0.338 0.00E+00 C1 R3HDM2 1.031695733 0.742 0.167 0.00E+00 C1 B3GNT7 1.031632795 0.549 0.088 0.00E+00 C1 MORF4L1 1.023176967 0.768 0.409 0.00E+00 C1 TMEM165 0.999912261 0.649 0.196 0.00E+00 C1 GALNT3 0.995165397 0.705 0.254 0.00E+00 C1 TFF1 0.962929536 0.96 0.591 0.00E+00 C1 LY6D 0.94328396 0.484 0.007 0.00E+00 C1 MIA 0.895355862 0.623 0.103 0.00E+00 C1 MDM2 0.864864363 0.615 0.089 0.00E+00 C1 YEATS4 0.864197638 0.585 0.128 0.00E+00 C1 CPM 0.831257805 -

Protein Kinase CK2 Potentiates Translation Efficiency By
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Biochimica et Biophysica Acta 1853 (2015) 1693–1701 Contents lists available at ScienceDirect Biochimica et Biophysica Acta journal homepage: www.elsevier.com/locate/bbamcr Protein kinase CK2 potentiates translation efficiency by phosphorylating eIF3j at Ser127 Christian Borgo a,b, Cinzia Franchin c,ValentinaSalizzatoa,b,LucaCesaroa,b, Giorgio Arrigoni c, Laura Matricardi d, Lorenzo A. Pinna a,b, Arianna Donella-Deana a,b,⁎ a Department of Biomedical Sciences, University of Padova, Via U. Bassi 58B, 35131 Padova, Italy b CNR Institute of NeuroSciences, University of Padova, Via U. Bassi 58B, 35131 Padova, Italy c Proteomic Center of Padova University, Via G. Orus B2, 35129 Padova, Italy d Venitian Institute of Oncology (IOV-IRCCS), Via Gattamelata 64, 35128 Padova, Italy article info abstract Article history: In eukaryotic protein synthesis the translation initiation factor 3 (eIF3) is a key player in the recruitment and Received 18 December 2014 assembly of the translation initiation machinery. Mammalian eIF3 consists of 13 subunits, including the loosely Received in revised form 17 March 2015 associated eIF3j subunit that plays a stabilizing role in the eIF3 complex formation and interaction with the Accepted 7 April 2015 40S ribosomal subunit. By means of both co-immunoprecipitation and mass spectrometry analyses we demon- Available online 15 April 2015 strate that the protein kinase CK2 interacts with and phosphorylates eIF3j at Ser127. Inhibition of CK2 activity by CX-4945 or down-regulation of the expression of CK2 catalytic subunit by siRNA cause the dissociation of Keywords: fi eIF3j phosphorylation j-subunit from the eIF3 complex as judged from glycerol gradient sedimentation.