Pituitary Gland Structure and Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

P21 Restrains Pituitary Tumor Growth
p21Cip1 restrains pituitary tumor growth Vera Chesnokovaa, Svetlana Zonisa, Kalman Kovacsb, Anat Ben-Shlomoa, Kolja Wawrowskya, Serguei Bannykhc and Shlomo Melmeda,1 Departments of aMedicine and cPathology, Cedars-Sinai Medical Center, Los Angeles, California 90048, bSt. Michael’s Hospital, Toronto, ON, Canada M5B 1W8 Edited by Wylie W. Vale, The Salk Institute for Biological Studies, La Jolla, CA, and approved September 15, 2008 (received for review May 16, 2008) As commonly encountered, pituitary adenomas are invariably benign. highlighting the securin requirement for the maintenance of chro- We therefore studied protective pituitary proliferative mechanisms. mosomal stability after genotoxic stress (24). Pituitary tumor transforming gene (Pttg) deletion results in pituitary Pituitary-directed transgenic Pttg overexpression results in focal p21 induction and abrogates tumor development in Rb؉/؊Pttg؊/؊ pituitary hyperplasia and small but functional adenoma formation mice. p21 disruption restores attenuated Rb؉/؊Pttg؊/؊ pituitary pro- (14). In contrast, mice lacking Pttg exhibit selective endocrine cell liferation rates and enables high penetrance of pituitary, but not hypoplasia (25). The permissive role of PTTG for pituitary ade- thyroid, tumor growth in triple mutant animals (88% of Rb؉/؊ and noma development was further confirmed by the observation that ϩ Ϫ -of Rb؉/؊Pttg؊/؊p21؊/؊ vs. 30% of Rb؉/؊Pttg؊/؊ mice developed Pttg deletion from the Rb / background results in p53/p21 senes 72% pituitary tumors, P < 0.001). p21 deletion also accelerated S-phase cence pathway activation, associated with pituitary hypoplasia and entry and enhanced transformation rates in triple mutant MEFs. attenuated adenoma formation (23, 26). Intranuclear p21 accumulates in Pttg-null aneuploid GH-secreting p21Cip/Kip (p21), a transcriptional target of p53, is induced in cells, and GH3 rat pituitary tumor cells overexpressing PTTG also response to a spectrum of cellular stresses and acts to constrain the exhibited increased levels of mRNA for both p21 (18-fold, P < 0.01) cell cycle. -

The Immuno-Neuroendocrine Interface
Amendment history: Erratum (December 2001) Series Introduction: The immuno-neuroendocrine interface Shlomo Melmed J Clin Invest. 2001;108(11):1563-1566. https://doi.org/10.1172/JCI14604. Perspective The CNS and the various endocrine organs are linked in a dynamic manner through networks of reciprocal interactions that allow for both long-term adaptation and short-term shifts in environmental conditions. These regulatory systems embrace the hypothalamus, the pituitary gland, and any of several target glands, including the adrenal gland and the thyroid. Because the anterior pituitary gland secretes at least six key hormones — adrenocorticotropin (ACTH), growth hormone (GH), prolactin (PRL), thyrotropin (TSH), and the gonadotropins follicle stimulating hormone and luteinizing hormone — such diverse parameters as cell integrity, stress responses, growth, development, reproduction, and energy homeostasis are all directly or indirectly under central control (1). In addition, largely because of the dominant role of the hypothalamo-pituitary-adrenal (HPA) axis, inflammation and immunity are also subject to neuroendocrine effects. The articles in this Perspective series explore some of the developmental, physiological, and molecular interactions occurring at the immuno-neuroendocrine interface. Control of pituitary function Three tiers of control subserve regulation of anterior pituitary trophic hormone secretion (2). First, the hypothalamus synthesizes and secretes releasing and inhibiting hormones, which traverse the hypothalamo-pituitary portal system and bind to specific anterior pituitary G protein−linked transmembrane […] Find the latest version: https://jci.me/14604/pdf PERSPECTIVE SERIES Neuro-immune interface Shlomo Melmed, Series Editor SERIES INTRODUCTION The immuno-neuroendocrine interface Shlomo Melmed Cedars-Sinai Research Institute, University of California Los Angeles, School of Medicine, Los Angeles, California 90048, USA. -

Human General Histology
135 اووم څپرکي انډوکراين سيستم (Endocrine system) (hormones) (target cells) (receptors) autonomic (sinusoids) ductless glands thyroid gland Pineal gland Hypophysis cerebri(pituitary glands) supra renal( adrenal glands) parathyroid glands 136 اووم څپرکي انډوکراين سيسټم islets cell corpora lutea interstitial tissue (placenta) GIT amines neurotransmitters amines neuromodulator 137 اووم څپرکي انډوکراين سيسټم APUD cells neuroendocrine system system adrenaline, (amino acid derivatives) thyroxin noradrenalin thyroid vasopressin encephalin (small peptides) releasing hormone(TRH) TSH(thyroid stimulating parathormone hormone) cortisol Testosterone estrogen (steroids) 5,12,3 (Hypophysis Cerebri) (brain) pituitary gland (stalk) Ventricle infundibulum stalk pituitary fossa sphenoid pineal hypothalamus body 138 اووم څپرکي انډوکراين سيسټم Hypophysis cerebri Pars pars anterior pars nervosa pars posterior intermediate hypothalamus infundibulum infundibulum stalk )Pars posterior neurohypophysis median eminence (tuber cinereum) infundibulum pars neurohypophysis median eminence pars intermediate pars distalis anterior Adenohypophysis infundibulum pars anterior pars tuberalis adenohypophysis 139 اووم څپرکي انډوکراين سيسټم Adenohypophysis pars intermediate pars anterior adenohypophyisis Pars anterior fenestrated sinusoids (cords) chromophil chromophobic acidophil chromophil basophils orange G eosin PAS-positive hematoxylline Beta cells basophil Alpha cells Acidophil basophils acidophil (dese cored vesicles) alpha Beta Histochemical 140 اووم څپرکي انډوکراين -

The Physiology of Growth by S
Postgrad Med J: first published as 10.1136/pgmj.26.298.417 on 1 August 1950. Downloaded from 417 THE PHYSIOLOGY OF GROWTH BY S. LEONARD SIMPSON, M.A., M.D., F.R.C.P. Consultant Endocrinologist, St. Mary's Hospital, etc.; Formerly Research Wor7Aer, Lister Institute Although clinical and experimental evidence postulated an antagonism between growth and had previously indicated that skeletal growth sex-stimulating hormones. Further observations depended upon the pituitary gland, it was not suggested that corpora lutea were formed before until I92I that Evans and Long, at the University ovulation occurred, and that his emulsions con- of California, demonstrated that a saline emulsion tained gonadotrophic luteinizing hormone. Al- made from the anterior pituitary lobe of fresh though Evans' original giant rats were apparently glands of cattle contained a growth hormone. The adipose, later work (Lee and Schaffer, 1934), hypophyses were thrown into 40 per cent. ethyl showed that there was a retention of nitrogen and alcohol, agitated with a glass rod, transferred to a relative excess of body protein. This was also two changes of sterile normal saline, and triturated true of the excess of tissue deposited in and with force and speed with ocean sand, diluted with around the abdomen. Evans stated in a recent saline, and decanted. The layers of sand, cell discussion in London that rats treated with pure fragments and opaque pink fluid permitted the growth hormone, unlike those in the initial latter to be decanted. This emulsion, when experiments with crude extracts, were not injected daily in doses of I to i ml. -

Histochemical Characterization and Functional Significance of the Hyperintense Signal on MR Images of the Posterior Pituitary
1079 Histochemical Characterization and Functional Significance of the Hyperintense Signal on MR Images of the Posterior Pituitary 1 John Kucharczyk ,2 MR imaging of the pituitary fossa characteristically shows a well-circumscribed area Walter Kucharczyk3 of high signal intensity in the posterior lobe on T1-weighted images. We used a Isabelle Berri,4 combination of high-field MR, electron microscopy, and histologic techniques in experi Jack de GroatS mental animals to determine whether the hyperintensity of the posterior lobe might be William Kelly6 functionally related to hormone neurosecretory processes, and to attempt to establish its chemical nature. Histologic sections of a dog's pituitary gland processed with lipid David Norman 1 1 specific markers showed intense staining in the posterior lobe but not in the anterior T. H. Newton lobe, thus documenting the location of fat in the posterior pituitary. Administration of vasoactive drugs known to influence vasopressin secretion to anesthetized cats pro duced changes in the volume of high-intensity signal in the posterior pituitary. Subse quent electron microscopy showed a significant increase in posterior lobe glial cell lipid droplets and neurosecretory granules in dehydration-stimulated cats. The data suggest that the pituitary hyperintensity represents intracellular lipid signal in the glial cell pituicytes of the posterior lobe or neurosecretory granules containing vasopressin. The volume of the signal may, in turn, reflect the functional state of hormonal release from the neurohypophysis. While CT and MR imaging can both be used to evaluate patients with suspected pituitary disease, high-field high-resolution MR imaging is increasingly the method of choice [1-3]. -

Quantitation of Corticotrophs in the Pars Distalis of Stress-Prone Swine Beverly Ann Bedford Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1-1-1976 Quantitation of corticotrophs in the pars distalis of stress-prone swine Beverly Ann Bedford Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Veterinary Anatomy Commons Recommended Citation Bedford, Beverly Ann, "Quantitation of corticotrophs in the pars distalis of stress-prone swine" (1976). Retrospective Theses and Dissertations. 17953. https://lib.dr.iastate.edu/rtd/17953 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Quantitation of corticotrophs in the pars distalis of stress-prone swine by Beverly Ann Bedford A Thesis Submitted to the Graduate Faculty in Partial Fulfillment of The Requirements for the Degr~e of MASTER OF SCIENCE Department: Veterinary Anatomy, Pharmacology and Physiology Major: Veterinary Anatomy ., Signatures have been redacted for privacy ' I Iowa State University Ames, Iowa 1976 ii :E5 ll I q7(p ,g3r TABLE OF CONTENTS c,J. Page INTRODUCTION 1 LITERATURE REVIEW 4 Pituitary Gland 4 General morphology 4 Development 5 Blood supply 7 Staining techniques 7 Pars distalis 13 . Pars tuberalis 25 ·pars intermedia 28 Process of secretion 36 Neurohypophysis 41 Porcine Stress Syndrome 45 MATERIALS AND MET!iODS' 52 RESULTS 56 DISCUSSION 64 SUMMARY AND CONCLUSIONS 72 BIBLIOGRAPHY 73 ACKNOWLEDGMENTS 85 APPENDIX 86 1111408 1 INTRODUCTION As early as 1953, there came reports (Ludvigsen, 1953; Briskey et al., 1959) of pale soft exudative (PSE) post-mortem porcine muscu- lature which later stimulated research into the mechanisms responsible for this condition. -
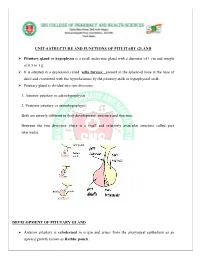
Unit-4 Structure and Functions of Pituitary Gland
UNIT-4 STRUCTURE AND FUNCTIONS OF PITUITARY GLAND Pituitary gland or hypophysis is a small endocrine gland with a diameter of 1 cm and weight of 0.5 to 1 g. It is situated in a depression called ‘sella turcica’, present in the sphenoid bone at the base of skull and connected with the hypothalamus by the pituitary stalk or hypophyseal stalk. Pituitary gland is divided into two divisions: 1. Anterior pituitary or adenohypophysis 2. Posterior pituitary or neurohypophysis. Both are entirely different in their development, structure and function. Between the two divisions, there is a small and relatively avascular structure called pars intermedia. DEVELOPMENT OF PITUTARY GLAND Anterior pituitary is ectodermal in origin and arises from the pharyngeal epithelium as an upward growth known as Rathke pouch. Posterior pituitary is neuroectodermal in origin and arises from hypothalamus as a downward diverticulum. Rathke pouch and the downward diverticulum from hypothalamus grow towards each other and meet in the midway between the roof of the buccal cavity and base of brain. There, the two structures lie close together. ANTERIOR PITUITARY OR ADENOHYPOPHYSIS Anterior pituitary is also known as the master gland because it regulates many other endocrine glands through its hormones. Anterior pituitary consists of three parts 1. Pars distalis 2. Pars tuberalis 3. Pars intermedia. Anterior pituitary has two types of cells 1. Chromophobe cells 2. Chromophil cells. Chromophobe Cells Chromophobe cells are not secretory in nature, but are the precursors of chromophil cells. Chromophil Cells On the basis of secretory nature chromophil cells are classified into five types: i. -

Endocrine System Hormonal Regulation Endocrine Glands
Endocrine system Hormonal regulation Endocrine glands • Glands w/o ducts • Secretory cells release their products, hormones, into the extracellular space and blood stream • Alternatively, the hormones may affect neighbor cells (paracrine) • Structure: • c.t. – capsule + septs • irregular clumps or cords of the cells • network of capillaries • fenestrated capillaries • sinusoids Hypophysis – pituitary gland Pituitary gland – anterior pituitary Pituitary gland – anterior pituitary Chromophil cells - Acidophilic cells (produce proteins) somatotrophs mammotrophs (or lactotrophs) - Basophilic cells (produce glycoproteins) thyrotrophs produce thyroid stimulating hormone (TSH or thyrotropin). gonadotrophs produce follicle stimulating hormone (FSH) and luteinizing hormone (LH) corticotrophs (or adrenocorticolipotrophs) Chromophobe cells Adenohypophysis Adenohypophysis oranž G + PAS Neurohypophysis x Adenohypophysis Neurohypophysis - structure Structure unmyelinated nerve fibres derived from neurosecretory cells of the supraoptic and paraventricular hypothalamic nuclei pituicytes /neuroglia/ Function Two hormones are oxytocin, which stimulates the contraction of smooth muscle cell in the uterus and participates in the milk ejection reflex, and antidiuretic hormone (ADH or vasopressin), which facilitates the concentration of urine in the kidneys and, thereby, the retention of water. Usually only the oval or round nuclei of the pituicytes are visible. Hypothalamic nerve fibres typically terminate close to capillaries. Scattered, large -

Khat-Induced Reproductive Dysfunction in Male Rabbits
KHAT-INDUCED REPRODUCTIVE DYSFUNCTION IN MALE RABBITS. NYONGESA ALBERT WAFULA, BVM (UON) DEPARTMENT OF VETERINARY ANATOMY AND PHYSIOLOGY UNIVERSITY OF NAIROBI A thesis submitted in partial fulfilment of the requirements for the award of the degree of Master of Science in Comparative Mammalian Physiology of the University of Nairobi. 2006. ii iii ACKNOWLEDGEMENTS Special thanks go to my supervors: Prof. Nilesh B. Patel, Prof. Emmanuel O. Wango and Dr. Daniel W. Onyango for their untiring efforts and willingness to assist me from start to the completion of this research project. Their constructive comments, patience and supervision throughout the project and write up of the thesis are appreciated. I appreciate the financial support from the University of Nairobi for granting me the scholarship to pursue this work. I also wish to thank WHO Special Programme for Research Development and Training in Human Reproduction for donating radioimmunoassay reagents without which this project would not have been successful. I also wish to thank the Chairman, Department of Zoology, University of Nairobi, for providing me with space, free use of animal house facilities and personnel. My other appreciation goes to H. Odongo and J. Winga for their technical assistance in haematological and hormonal assays procedures; J.M Gachoka for his technical assistance in histology and photography and A. Tangai for his excellent computer skills. My sincere gratitude goes to animal handlers F.K Kamau, J.K Samoei and P. Simiyu for their untiring efforts in looking after the animals. Finally, my special thanks to my friends Dr. Egesa, Dr. Njogu, Dr. Kavoi, Dr. Kisipan, S. -

Suprarenocortical Syndrome and Pituitary Basophilism
University of Nebraska Medical Center DigitalCommons@UNMC MD Theses Special Collections 5-1-1935 Suprarenocortical syndrome and pituitary basophilism Leonard H. Barber University of Nebraska Medical Center This manuscript is historical in nature and may not reflect current medical research and practice. Search PubMed for current research. Follow this and additional works at: https://digitalcommons.unmc.edu/mdtheses Part of the Medical Education Commons Recommended Citation Barber, Leonard H., "Suprarenocortical syndrome and pituitary basophilism" (1935). MD Theses. 630. https://digitalcommons.unmc.edu/mdtheses/630 This Thesis is brought to you for free and open access by the Special Collections at DigitalCommons@UNMC. It has been accepted for inclusion in MD Theses by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. SUPRARENOCORTIOAL SYNDROME AND P·ITUITARY BASOPHILISM - L!X>NARD HOBBS BARBER SENIOR THESIS UNIVERSITY OF NEBRASKA· COLLEGE OF MEDICINE. 1935 A.Introduction 1 B.Anatomy of the Hypophysis and Adrenal 6 C.Normal Physiology of the Pituitary 8 D. The Pituitary Adenomas 16 E.Basophilic Syndrome of the Pituitary 21 F.Case Histories 23 1. Discussion of Cases 38 G.Summary and Conclusions 52 H. Bibliography 54 TABLE OF CONTENTS 480672 -1- INTRODUCTION From va.rious sources in recent years new facts have been unearthed both in clinic and laboratory which have thrown light on many heretofore obscure activities of the pituitary gland and adrenal gland. The existance of what we now sneak of as an internal secretion was -perha:os first experimentally demonstrated by Berthold's studies in 1849 on transplantation of the cock's testis. -

Cell Population Characterization and Discovery Using Single-Cell Technologies in Endocrine Systems
65 2 Journal of Molecular LYM Cheung and K Rizzoti Single cell technologies in 65:2 R35–R51 Endocrinology endocrine systems REVIEW Cell population characterization and discovery using single-cell technologies in endocrine systems Leonard Y M Cheung 1 and Karine Rizzoti 2 1Department of Human Genetics, University of Michigan, Michigan, Ann Arbor, USA 2Laboratory of Stem Cell Biology and Developmental Genetics, The Francis Crick Institute, London, UK Correspondence should be addressed to K Rizzoti: [email protected] Abstract In the last 15 years, single-cell technologies have become robust and indispensable Key Words tools to investigate cell heterogeneity. Beyond transcriptomic, genomic and epigenome f technology analyses, technologies are constantly evolving, in particular toward multi-omics, where f single cell analyses of different source materials from a single cell are combined, and spatial f microfluidics transcriptomics, where resolution of cellular heterogeneity can be detected in situ. While f multi-omics some of these techniques are still being optimized, single-cell RNAseq has commonly f transcriptome been used because the examination of transcriptomes allows characterization of f endocrine organs cell identity and, therefore, unravel previously uncharacterized diversity within cell populations. Most endocrine organs have now been investigated using this technique, and this has given new insights into organ embryonic development, characterization of rare cell types, and disease mechanisms. Here, we highlight recent studies, particularly on the hypothalamus and pituitary, and examine recent findings on the pancreas and Journal of Molecular reproductive organs where many single-cell experiments have been performed. Endocrinology (2020) 65, R35–R51 Introduction Single-cell technologies have become an essential soon be analyzed at the single-cell level (Palii et al. -

REVIEW Diagnosis of Pheochromocytoma
Clin. Lab. 2002;48:5-18 ÓCopyright REVIEW Diagnosis of Pheochromocytoma TOMAS LENZ, JAN GOSSMANN, KARL-LUDWIG SCHULTE*, LOTHAR SALEWSKI** AND HELMUT GEIGER Medical Clinic IV, Division of Nephrology, Johann Wolfgang Goethe University, Frankfurt am Main, Germany *Medical Clinic, Königin Elisabeth Herzberge Hospital, Berlin, Germany **Immnno Biological Laboratories, Hamburg, Germany SUMMARY The purpose of this article is to give an overview on recent advances in the diagnosis, localization by imaging and treatment of pheochromocytoma. Pheochromocytoma is a mostly benign tumor (malignancy rate 10 - 15%) which arises from chromaffin cells with excessive catecholamine production and secretion. Most tumors are localized in the adrenals but 15 - 18% of the lesions are found extraadrenally (paragangliomas). Pheochromocytoma is a rare form of secondary hypertension; it can also be found as a feature of familial disease (e.g. von Hippel-Lindau disease, MEN type II) due to genetic mutations of several genes that have been identified recently. In familial pheochromocytoma molecular genetic analysis has improved the diagnostic modalities. In such patients the tumor can occur bilaterally and patients often remain normotensive until the tumor produces sufficient catecholamines to have hemodynamic effects. The extreme importance of recognizing this tumor is evident from the fact that it can be successfully removed in about 90% of the cases, whereas if unrecognized the tumor poses great risk of death or devastating complications. Diagnostic screening includes measurement of catecholamines and their metabolites (metanephrines) in plasma and/or urine. Furthermore, pharmacological testing (e.g. clonidine suppression test) may be indicated in patients with moderately elevated catecholamines or when the diagnosis is still uncertain.