The Chamois (Rupicapra Cf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Capture, Restraint and Transport Stress in Southern Chamois (Rupicapra Pyrenaica)
Capture,Capture, restraintrestraint andand transporttransport stressstress ininin SouthernSouthern chamoischamois ((RupicapraRupicapra pyrenaicapyrenaica)) ModulationModulation withwith acepromazineacepromazine andand evaluationevaluation usingusingusing physiologicalphysiologicalphysiological parametersparametersparameters JorgeJorgeJorge RamónRamónRamón LópezLópezLópez OlveraOlveraOlvera 200420042004 Capture, restraint and transport stress in Southern chamois (Rupicapra pyrenaica) Modulation with acepromazine and evaluation using physiological parameters Jorge Ramón López Olvera Bellaterra 2004 Esta tesis doctoral fue realizada gracias a la financiación de la Comisión Interministerial de Ciencia y Tecnología (proyecto CICYT AGF99- 0763-C02) y a una beca predoctoral de Formación de Investigadores de la Universidad Autónoma de Barcelona, y contó con el apoyo del Departament de Medi Ambient de la Generalitat de Catalunya. Los Doctores SANTIAGO LAVÍN GONZÁLEZ e IGNASI MARCO SÁNCHEZ, Catedrático de Universidad y Profesor Titular del Área de Conocimiento de Medicina y Cirugía Animal de la Facultad de Veterinaria de la Universidad Autónoma de Barcelona, respectivamente, CERTIFICAN: Que la memoria titulada ‘Capture, restraint and transport stress in Southern chamois (Rupicapra pyrenaica). Modulation with acepromazine and evaluation using physiological parameters’, presentada por el licenciado Don JORGE R. LÓPEZ OLVERA para la obtención del grado de Doctor en Veterinaria, se ha realizado bajo nuestra dirección y, considerándola satisfactoriamente -

Contributions to the 12Th Conference of the European Wildlife Disease Association (EWDA) August 27Th – 31St, 2016, Berlin
12th Conference of the European Wildlife Disease Association (EWDA), Berlin 2016 Contributions to the 12th Conference of the European Wildlife Disease Association (EWDA) August 27th – 31st, 2016, Berlin, Germany Edited by Anke Schumann, Gudrun Wibbelt, Alex D. Greenwood, Heribert Hofer Organised by Leibniz Institute for Zoo and Wildlife Research (IZW) Alfred-Kowalke-Straße 17 10315 Berlin Germany www.izw-berlin.de and the European Wildlife Disease Association (EWDA) https://sites.google.com/site/ewdawebsite/ & ISBN 978-3-9815637-3-3 12th Conference of the European Wildlife Disease Association (EWDA), Berlin 2016 Published by Leibniz Institute for Zoo and Wildlife Research (IZW) Alfred-Kowalke-Str. 17, 10315 Berlin (Friedrichsfelde) PO Box 700430, 10324 Berlin, Germany Supported by Deutsche Forschungsgemeinschaft (DFG) [German Research Foundation] Kennedyallee 40, 53175 Bonn, Germany Printed on Forest Stewardship Council certified paper All rights reserved, particularly those for translation into other languages. It is not permitted to reproduce any part of this book by photocopy, microfilm, internet or any other means without written permission of the IZW. The use of product names, trade names or other registered entities in this book does not justify the assumption that these can be freely used by everyone. They may represent registered trademarks or other legal entities even if they are not marked as such. Processing of abstracts: Anke Schumann, Gudrun Wibbelt Setting and layout: Anke Schumann, Gudrun Wibbelt Cover: Diego Romero, Steven Seet, Gudrun Wibbelt Word cloud: ©Tagul.com Printing: Spree Druck Berlin GmbH www.spreedruck.de Order: Leibniz Institute for Zoo and Wildlife Research (IZW) Forschungsverbund Berlin e.V. PO Box 700430, 10324 Berlin, Germany [email protected] www.izw-berlin.de 12th Conference of the European Wildlife Disease Association (EWDA), Berlin 2016 CONTENTS Foreword ................................................................................................................. -

Anaplasma Phagocytophilum and Babesia Species Of
pathogens Article Anaplasma phagocytophilum and Babesia Species of Sympatric Roe Deer (Capreolus capreolus), Fallow Deer (Dama dama), Sika Deer (Cervus nippon) and Red Deer (Cervus elaphus) in Germany Cornelia Silaghi 1,2,*, Julia Fröhlich 1, Hubert Reindl 3, Dietmar Hamel 4 and Steffen Rehbein 4 1 Institute of Comparative Tropical Medicine and Parasitology, Ludwig-Maximilians-Universität München, Leopoldstr. 5, 80802 Munich, Germany; [email protected] 2 Institute of Infectology, Friedrich-Loeffler-Institut, Südufer 10, 17493 Greifswald Insel Riems, Germany 3 Tierärztliche Fachpraxis für Kleintiere, Schießtrath 12, 92709 Moosbach, Germany; [email protected] 4 Boehringer Ingelheim Vetmedica GmbH, Kathrinenhof Research Center, Walchenseestr. 8-12, 83101 Rohrdorf, Germany; [email protected] (D.H.); steff[email protected] (S.R.) * Correspondence: cornelia.silaghi@fli.de; Tel.: +49-0-383-5171-172 Received: 15 October 2020; Accepted: 18 November 2020; Published: 20 November 2020 Abstract: (1) Background: Wild cervids play an important role in transmission cycles of tick-borne pathogens; however, investigations of tick-borne pathogens in sika deer in Germany are lacking. (2) Methods: Spleen tissue of 74 sympatric wild cervids (30 roe deer, 7 fallow deer, 22 sika deer, 15 red deer) and of 27 red deer from a farm from southeastern Germany were analyzed by molecular methods for the presence of Anaplasma phagocytophilum and Babesia species. (3) Results: Anaplasma phagocytophilum and Babesia DNA was demonstrated in 90.5% and 47.3% of the 74 combined wild cervids and 14.8% and 18.5% of the farmed deer, respectively. Twelve 16S rRNA variants of A. phagocytophilum were delineated. -
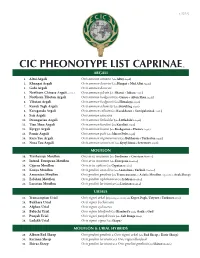
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26. -

Spatial Patterns of Mitochondrial and Nuclear Gene Pools in Chamois (Rupicapra R
Heredity (2003) 91, 125–135 & 2003 Nature Publishing Group All rights reserved 0018-067X/03 $25.00 www.nature.com/hdy Spatial patterns of mitochondrial and nuclear gene pools in chamois (Rupicapra r. rupicapra) from the Eastern Alps H Schaschl, D Kaulfus, S Hammer1 and F Suchentrunk Research Institute of Wildlife Ecology, Veterinary Medicine University of Vienna, Savoyenstrasse 1, A-1160 Vienna, Austria We have assessed the variability of maternally (mtDNA) and variability as a result of immigration of chamois from different biparentally (allozymes) inherited genes of 443 chamois Pleistocene refugia surrounding the Alps after the withdrawal (Rupicapra r. rupicapra) from 19 regional samples in the of glaciers, rather than from topographic barriers to gene Eastern Alps, to estimate the degree and patterns of spatial flow, such as Alpine valleys, extended glaciers or woodlands. gene pool differentiation, and their possible causes. Based However, this striking geographical structuring of the on a total mtDNA-RFLP approach with 16 hexanucleotide- maternal genome was not paralleled by allelic variation at recognizing restriction endonucleases, we found marked 33 allozyme loci, which were used as nuclear DNA markers. substructuring of the maternal gene pool into four phylogeo- Wright’s hierarchical F-statistics revealed that only p0.45% of graphic groups. A hierarchical AMOVA revealed that 67.09% the explained allozymic diversity was because of partitioning of the variance was partitioned among these four mtDNA- among the four mtDNA-phylogroups. We conclude that this phylogroups, whereas only 8.04% were because of partition- discordance of spatial patterns of nuclear and mtDNA gene ing among regional samples within the populations, and pools results from a phylogeographic background and sex- 24.86% due to partitioning among individuals within regional specific dispersal, with higher levels of philopatry in females. -

Temporal Variation in Foraging Activity and Grouping Patterns in a Mountain-Dwelling Herbivore Environmental and Endogenous
Behavioural Processes 167 (2019) 103909 Contents lists available at ScienceDirect Behavioural Processes journal homepage: www.elsevier.com/locate/behavproc Temporal variation in foraging activity and grouping patterns in a mountain-dwelling herbivore: Environmental and endogenous drivers T ⁎ Niccolò Fattorinia, , Claudia Brunettia, Carolina Baruzzia, Gianpasquale Chiatanteb, Sandro Lovaria,c, Francesco Ferrettia a Department of Life Sciences, University of Siena. Via P.A. Mattioli 4, 53100 Siena, Italy b Department of Earth and Environmental Sciences, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy c Maremma Natural History Museum, Strada Corsini 5, 58100 Grosseto, Italy ARTICLE INFO ABSTRACT Keywords: In temperate ecosystems, seasonality influences animal behaviour. Food availability, weather, photoperiod and Seasonality endogenous factors relevant to the biological cycle of individuals have been shown as major drivers of temporal Foraging Activity changes in activity rhythms and group size/structure of herbivorous species. We evaluated how diurnal female Group size foraging activity and grouping patterns of a mountain herbivore, the Apennine chamois Rupicapra pyrenaica Rupicapra ornata, varied during a decreasing gradient of pasture availability along the summer-autumn progression Chamois (July–October), a crucial period for the life cycle of mountain ungulates. Mountain herbivores Females increased diurnal foraging activity, possibly because of constrains elicited by variation in environ- mental factors. Size of mixed groups did not vary, in contrast with the hypothesis that groups should be smaller when pasture availability is lower. Proportion of females in groups increased, possibly suggesting that they concentrated on patchily distributed nutritious forbs. Occurrence of yearlings in groups decreased, which may have depended on dispersal of chamois in this age class. -

Balkan Chamois (Rupicapra Rupicapra Balcanica) Avoids Roads, Settlements, and Hunting Grounds: an Ecological Overview from Timfi Mountain, Greece
diversity Article Balkan Chamois (Rupicapra rupicapra balcanica) Avoids Roads, Settlements, and Hunting Grounds: An Ecological Overview from Timfi Mountain, Greece Vassiliki Kati 1,* , Christina Kassara 1, Dimitrios Vassilakis 2 and Haritakis Papaioannou 1,3 1 Department of Biological Applications & Technology, University of Ioannina, University Campus, 45500 Ioannina, Greece; [email protected] (C.K.); [email protected] (H.P.) 2 Hellenic Republic, Decentralized Administration of Macedonia–Thrace, Forestry Service of Soufli, Ermou 6, 68400 Soufli, Greece; [email protected] 3 Pindos Perivallontiki Non-Profit Organization, Metsovou 12, Ioannina 45221, Greece * Correspondence: [email protected]; Tel.: +30-265-100-7439 Received: 12 March 2020; Accepted: 24 March 2020; Published: 27 March 2020 Abstract: Balkan chamois (Rupicapra rupicapra balcanica) is a protected species with an Inadequate-Bad (U2) conservation status in Greece. Our study explores its seasonal range use pattern, demography and habitat selection in a site of the Natura 2000 network, Timfi Mountain. To this aim, we examined 1168 observations obtained from six seasonal surveys (2002: four seasons, 2014 and 2017: autumn) and performed an ecological-niche factor analysis (ENFA), using 16 environmental and human-disturbance variables. The species had an annual range of 6491 ha (25% of the study area), followed the typical range-use pattern, and presented the minimum core area during the rutting season (autumn). Timfi Mt hosted 469 individuals in 2017 (the largest population in Greece), increasing by 3.55 times since 2002. The species selected higher altitudes during summer and autumn, pinewoods over broad-leaved woods as winter grounds, and it avoided south-facing slopes. -

Diversity and Evolution of the Mhc-DRB1 Gene in the Two Endemic Iberian Subspecies of Pyrenean Chamois, Rupicapra Pyrenaica
Heredity (2007) 99, 406–413 & 2007 Nature Publishing Group All rights reserved 0018-067X/07 $30.00 www.nature.com/hdy ORIGINAL ARTICLE Diversity and evolution of the Mhc-DRB1 gene in the two endemic Iberian subspecies of Pyrenean chamois, Rupicapra pyrenaica J Alvarez-Busto1, K Garcı´a-Etxebarria1, J Herrero2,3, I Garin4 and BM Jugo1 1Genetika, Antropologia Fisikoa eta Animali Fisiologia Saila, Zientzia eta Teknologia Fakultatea, Euskal Herriko Unibertsitatea (UPV/EHU), Bilbao, Spain; 2Departamento de Ecologı´a, Universidad de Alcala´ de Henares, Alcala´ de Henares, Spain; 3EGA Wildlife Consultants, Zaragoza, Spain and 4Zoologı´a eta Animali Zelulen Biologia Saila, Zientzia eta Teknologia Fakultatea, Euskal Herriko Unibertsitatea (UPV/EHU), Bilbao, Spain Major histocompatibility complex class II locus DRB variation recent parasitic infections by sarcoptic mange. A phyloge- was investigated by single-strand conformation polymorph- netic analysis of both Pyrenean chamois and DRB alleles ism analysis and sequence analysis in the two subspecies of from 10 different caprinid species revealed that the chamois Pyrenean chamois (Rupicapra pyrenaica) endemic to the alleles form two monophyletic groups. In comparison with Iberian Peninsula. Low levels of genetic variation were other Caprinae DRB sequences, the Rupicapra alleles detected in both subspecies, with seven different alleles in R. displayed a species-specific clustering that reflects a large p. pyrenaica and only three in the R. p. parva. After applying temporal divergence of the chamois from other caprinids, as the rarefaction method to cope with the differences in sample well as a possible difference in the selective environment for size, the low allele number of parva was highlighted. -

List of 28 Orders, 129 Families, 598 Genera and 1121 Species in Mammal Images Library 31 December 2013
What the American Society of Mammalogists has in the images library LIST OF 28 ORDERS, 129 FAMILIES, 598 GENERA AND 1121 SPECIES IN MAMMAL IMAGES LIBRARY 31 DECEMBER 2013 AFROSORICIDA (5 genera, 5 species) – golden moles and tenrecs CHRYSOCHLORIDAE - golden moles Chrysospalax villosus - Rough-haired Golden Mole TENRECIDAE - tenrecs 1. Echinops telfairi - Lesser Hedgehog Tenrec 2. Hemicentetes semispinosus – Lowland Streaked Tenrec 3. Microgale dobsoni - Dobson’s Shrew Tenrec 4. Tenrec ecaudatus – Tailless Tenrec ARTIODACTYLA (83 genera, 142 species) – paraxonic (mostly even-toed) ungulates ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BOVIDAE (46 genera) - cattle, sheep, goats, and antelopes 1. Addax nasomaculatus - Addax 2. Aepyceros melampus - Impala 3. Alcelaphus buselaphus - Hartebeest 4. Alcelaphus caama – Red Hartebeest 5. Ammotragus lervia - Barbary Sheep 6. Antidorcas marsupialis - Springbok 7. Antilope cervicapra – Blackbuck 8. Beatragus hunter – Hunter’s Hartebeest 9. Bison bison - American Bison 10. Bison bonasus - European Bison 11. Bos frontalis - Gaur 12. Bos javanicus - Banteng 13. Bos taurus -Auroch 14. Boselaphus tragocamelus - Nilgai 15. Bubalus bubalis - Water Buffalo 16. Bubalus depressicornis - Anoa 17. Bubalus quarlesi - Mountain Anoa 18. Budorcas taxicolor - Takin 19. Capra caucasica - Tur 20. Capra falconeri - Markhor 21. Capra hircus - Goat 22. Capra nubiana – Nubian Ibex 23. Capra pyrenaica – Spanish Ibex 24. Capricornis crispus – Japanese Serow 25. Cephalophus jentinki - Jentink's Duiker 26. Cephalophus natalensis – Red Duiker 1 What the American Society of Mammalogists has in the images library 27. Cephalophus niger – Black Duiker 28. Cephalophus rufilatus – Red-flanked Duiker 29. Cephalophus silvicultor - Yellow-backed Duiker 30. Cephalophus zebra - Zebra Duiker 31. Connochaetes gnou - Black Wildebeest 32. Connochaetes taurinus - Blue Wildebeest 33. Damaliscus korrigum – Topi 34. -

Allozyme Divergence and Phylogenetic Relationships Among Capra, Ovis and Rupicapra (Artyodactyla, Bovidae)
Heredity'S? (1991) 281—296 Received 28 November 7990 Genetical Society of Great Britain Allozyme divergence and phylogenetic relationships among Capra, Ovis and Rupicapra (Artyodactyla, Bovidae) E. RANOI,* G. FUSCO,* A. LORENZINI,* S. TOSO & G. TOSIt *g7ftj Nez/c nate di 9/clog/a del/a Selvaggine, Via Ca Fornacetta, 9 Ozzano dell'EmiIia (Bo) Italy, and j-Dipartimento di B/Wag/a, Universitâ diM/lana, Via Ce/ar/a, 3 Mi/epa,Italy Geneticdivergence and phylogenetic relationships between the chamois (Rupicaprini, Rupkapra rupicapra rupicapra) and three species of the Caprini (Capra aegagrus hircus, Capra ibex ibex and Ovis amrnon musUnon) have been studied by multilocus protein electrophoresis. Dendrograms have been constructed both with distance and parsimony methods. Goat, sheep and chamois pair- wise genetic distances had very similar values, All the topologies showed that Capra, Ovis and Rupicapra originate from the same internode, suggesting the hypothesis of a common, and almost contemporaneous, ancestor. The estimated divergence times among the three genera ranged from 5.28 to 7.08 Myr. These findings suggest the need to reconsider the evolutionary relationships in the Caprinae. Keywords:allozymes,Caprinae, electrophoresis phylogenetic trees. caprid lineage since the lower or middle Miocene. Introduction ShaDer (1977) agrees with the outline given by Thenius Theevolutionary relationships of the subfamily & Hofer (1960) supporting the idea of a more recent Caprinae (Artyodactyla, Bovidae; Corbet, 1978) have origin of the Caprini, and in particular of a Pliocenic been discussed by Geist (197!) within the framework splitting of Ovis and Capra. In Geist's (1971) opinion of his dispersal theory of Ice Age mammal evolution. -

Infectious Keratoconjunctivitis at the Wildlife-Livestock Interface from Mountain Systems: Dynamics, Functional Roles and Host-Mycoplasma Interactions
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Infectious keratoconjunctivitis at the wildlife‐livestock interface from mountain systems Dynamics, functional roles and host‐ mycoplasma interactions Xavier Fernández Aguilar PHD Thesis Infectious keratoconjunctivitis at the wildlife-livestock interface from mountain systems: dynamics, functional roles and host-mycoplasma interactions Xavier Fernández Aguilar Directors Jorge Ramón López Olvera Óscar Cabezón Ponsoda Tesi Doctoral Departament de Medicina i Cirurgia Animals Facultat de Vetarinària Universitat Autònoma de Barcelona 2017 Drawings and pictures of this thesis are from the author except for: page 31, top: Radomir Jakubowski; page 37, top: Fernando Mostacero; page 48; figure 2.11 C: Arturo Galvez ; page 49, figure 2.12 C: Patrick Deleury, D: Santiago Lavín; page 63, figure 2.15 A, C: Etienne Florence and B, D: Jean Paul Crampe and E: Benoît Dandonneau; page 105, figure 4.2.2: Luca Rossi; page 133, figure 4.3.6: -

First Description of Onchocerca Jakutensis (Nematoda: Filarioidea) in Red Deer (Cervus Elaphus) in Switzerland
International Journal for Parasitology: Parasites and Wildlife 5 (2016) 192e197 Contents lists available at ScienceDirect International Journal for Parasitology: Parasites and Wildlife journal homepage: www.elsevier.com/locate/ijppaw First description of Onchocerca jakutensis (Nematoda: Filarioidea) in red deer (Cervus elaphus) in Switzerland * Felix Bosch a, Ralph Manzanell a, Alexander Mathis b, a Bündner Kantonsschule, Chur, Switzerland b Institute of Parasitology, Vetsuisse Faculty, University of Zürich, Zürich, Switzerland article info abstract Article history: Twenty-seven species of the genus Onchocerca (Nematoda; Filarioidea) can cause a vector-borne parasitic Received 10 March 2016 disease called onchocercosis. Most Onchocerca species infect wild and domestic ungulates or the dog, and Received in revised form one species causes river blindness in humans mainly in tropical Africa. The European red deer (Cervus e. 8 June 2016 elaphus) is host to four species, which are transmitted by blackflies (simuliids) or biting midges (cera- Accepted 10 June 2016 topogonids). Two species, Onchocerca flexuosa and Onchocerca jakutensis, produce subcutaneous nodules, whereas Onchocerca skrjabini and Onchocerca garmsi live free in the hypodermal serous membranes. Keywords: During the hunting season, September 2013, red deer (n ¼ 25), roe deer (Capreolus c. capreolus,n¼ 6) Onchocerca jakutensis ¼ Morphology and chamois (Rupicapra r. rupicapra,n 7), all shot in the Grisons Region (Switzerland) were investi- Cervus elaphus gated for the presence of subcutaneous nodules which were enzymatically digested, and the contained Red deer Onchocerca worms were identified to species by light and scanning electron microscopy as well as by Subcutaneous nodule PCR/sequencing. In addition, microfilariae from skin samples were collected and genetically character- Switzerland ized.