Supplemental Data FOXP2 Targets Show Evidence of Positive
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Reconstruction of the global neural crest gene regulatory network in vivo Ruth M Williams1, Ivan Candido-Ferreira1, Emmanouela Repapi2, Daria Gavriouchkina1,4, Upeka Senanayake1, Jelena Telenius2,3, Stephen Taylor2, Jim Hughes2,3, and Tatjana Sauka-Spengler1,∗ Supplemental Material ∗Lead and corresponding author: Tatjana Sauka-Spengler ([email protected]) 1University of Oxford, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, Oxford, OX3 9DS, UK 2University of Oxford, MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 3University of Oxford, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 4Present Address: Okinawa Institute of Science and Technology, Molecular Genetics Unit, Onna, 904-0495, Japan A 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R1_5-6ss) log2(R1_8-10ss) log2(R1_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.92 0 r=0.99 0 r=0.96 0 r=0.99 0 r=0.96 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R2_non-NC) log2(R2_5-6ss) log2(R3_5-6ss) log2(R2_8-10ss) log2(R3_8-10ss) 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R2_5-6ss) log2(R1_8-10ss) log2(R2_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.94 0 r=0.96 0 r=0.95 0 r=0.96 0 r=0.95 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R3_non-NC) log2(R4_5-6ss) log2(R3_5-6ss) log2(R4_8-10ss) log2(R3_8-10ss) -

Itcs) in the Mouse Amygdala of Tshz1 Mutants Correlates with Fear, Depression, and Social Interaction Phenotypes
1160 • The Journal of Neuroscience, January 31, 2018 • 38(5):1160–1177 Development/Plasticity/Repair Loss of Intercalated Cells (ITCs) in the Mouse Amygdala of Tshz1 Mutants Correlates with Fear, Depression, and Social Interaction Phenotypes X Jeffrey Kuerbitz,1 Melinda Arnett,5 Sarah Ehrman,1 XMichael T. Williams,3 XCharles V. Vorhees,3 X Simon E. Fisher,6,7 Alistair N. Garratt,8 XLouis J. Muglia,5 Ronald R. Waclaw,1,4 and XKenneth Campbell1,2 Divisions of 1Developmental Biology, 2Neurosurgery, 3Neurology, 4Experimental Hematology and Cancer Biology, 5Center for Prevention of Preterm Birth, Perinatal Institute, Cincinnati Children’s Hospital Medical Center, University of Cincinnati College of Medicine, Cincinnati, OH 45229, 6Language and Genetics Department, Max Planck Institute for Psycholinguistics, 6500 AH Nijmegen, The Netherlands, 7Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, The Netherlands, and 8Institute of Cell Biology and Neurobiology, Center for Anatomy, Charite´ University Hospital Berlin, 10117 Berlin, Germany The intercalated cells (ITCs) of the amygdala have been shown to be critical regulatory components of amygdalar circuits, which control appropriate fear responses. Despite this, the molecular processes guiding ITC development remain poorly understood. Here we establish the zinc finger transcription factor Tshz1 as a marker of ITCs during their migration from the dorsal lateral ganglionic eminence through maturity. Using germline and conditional knock-out (cKO) mouse models, we show that Tshz1 is required for the proper migration and differentiation of ITCs. In the absence of Tshz1, migrating ITC precursors fail to settle in their stereotypical locations encapsulating the lateral amygdala and BLA. Furthermore, they display reductions in the ITC marker Foxp2 and ectopic persistence of the dorsal lateral ganglionic eminence marker Sp8. -

Mediator of DNA Damage Checkpoint 1 (MDC1) Is a Novel Estrogen Receptor Co-Regulator in Invasive 6 Lobular Carcinoma of the Breast 7 8 Evelyn K
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.16.423142; this version posted December 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Running Title: MDC1 co-regulates ER in ILC 2 3 Research article 4 5 Mediator of DNA damage checkpoint 1 (MDC1) is a novel estrogen receptor co-regulator in invasive 6 lobular carcinoma of the breast 7 8 Evelyn K. Bordeaux1+, Joseph L. Sottnik1+, Sanjana Mehrotra1, Sarah E. Ferrara2, Andrew E. Goodspeed2,3, James 9 C. Costello2,3, Matthew J. Sikora1 10 11 +EKB and JLS contributed equally to this project. 12 13 Affiliations 14 1Dept. of Pathology, University of Colorado Anschutz Medical Campus 15 2Biostatistics and Bioinformatics Shared Resource, University of Colorado Comprehensive Cancer Center 16 3Dept. of Pharmacology, University of Colorado Anschutz Medical Campus 17 18 Corresponding author 19 Matthew J. Sikora, PhD.; Mail Stop 8104, Research Complex 1 South, Room 5117, 12801 E. 17th Ave.; Aurora, 20 CO 80045. Tel: (303)724-4301; Fax: (303)724-3712; email: [email protected]. Twitter: 21 @mjsikora 22 23 Authors' contributions 24 MJS conceived of the project. MJS, EKB, and JLS designed and performed experiments. JLS developed models 25 for the project. EKB, JLS, SM, and AEG contributed to data analysis and interpretation. SEF, AEG, and JCC 26 developed and performed informatics analyses. MJS wrote the draft manuscript; all authors read and revised the 27 manuscript and have read and approved of this version of the manuscript. -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -

Open Dogan Phdthesis Final.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science ELUCIDATING BIOLOGICAL FUNCTION OF GENOMIC DNA WITH ROBUST SIGNALS OF BIOCHEMICAL ACTIVITY: INTEGRATIVE GENOME-WIDE STUDIES OF ENHANCERS A Dissertation in Biochemistry, Microbiology and Molecular Biology by Nergiz Dogan © 2014 Nergiz Dogan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 ii The dissertation of Nergiz Dogan was reviewed and approved* by the following: Ross C. Hardison T. Ming Chu Professor of Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee David S. Gilmour Professor of Molecular and Cell Biology Anton Nekrutenko Professor of Biochemistry and Molecular Biology Robert F. Paulson Professor of Veterinary and Biomedical Sciences Philip Reno Assistant Professor of Antropology Scott B. Selleck Professor and Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School iii ABSTRACT Genome-wide measurements of epigenetic features such as histone modifications, occupancy by transcription factors and coactivators provide the opportunity to understand more globally how genes are regulated. While much effort is being put into integrating the marks from various combinations of features, the contribution of each feature to accuracy of enhancer prediction is not known. We began with predictions of 4,915 candidate erythroid enhancers based on genomic occupancy by TAL1, a key hematopoietic transcription factor that is strongly associated with gene induction in erythroid cells. Seventy of these DNA segments occupied by TAL1 (TAL1 OSs) were tested by transient transfections of cultured hematopoietic cells, and 56% of these were active as enhancers. Sixty-six TAL1 OSs were evaluated in transgenic mouse embryos, and 65% of these were active enhancers in various tissues. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
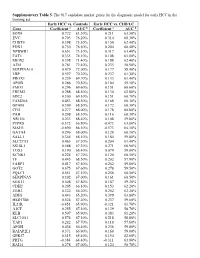
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

An Unbiased Reconstruction of the T Helper Cell Type 2 Differentiation Network
bioRxiv preprint doi: https://doi.org/10.1101/196022; this version posted October 4, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. An unbiased reconstruction of the T helper cell type 2 differentiation network 1,3 1 1 1 1 Authors: Johan Henriksson , Xi Chen , Tomás Gomes , Kerstin Meyer , Ricardo Miragaia , 4 1 4 1 1,2,* Ubaid Ullah , Jhuma Pramanik , Riita Lahesmaa , Kosuke Yusa , Sarah A Teichmann Affiliations: 1 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, United Kingdom 2 EMBL-European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SD, United Kingdom 3 Karolinska Institutet, Department. of Biosciences and Nutrition, Hälsovägen 7, Novum, SE-141 83, Huddinge, Sweden 4 Turku Centre for Biotechnology, Tykistokatu 6 FI-20520, Turku, Finland *To whom correspondence should be addressed: [email protected] Tomas: [email protected] Ricardo: [email protected] Ubaid Ullah: [email protected] Jhuma: [email protected] -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Identification of Key Genes and Pathways Involved in Response To
Deng et al. Biol Res (2018) 51:25 https://doi.org/10.1186/s40659-018-0174-7 Biological Research RESEARCH ARTICLE Open Access Identifcation of key genes and pathways involved in response to pain in goat and sheep by transcriptome sequencing Xiuling Deng1,2†, Dong Wang3†, Shenyuan Wang1, Haisheng Wang2 and Huanmin Zhou1* Abstract Purpose: This aim of this study was to investigate the key genes and pathways involved in the response to pain in goat and sheep by transcriptome sequencing. Methods: Chronic pain was induced with the injection of the complete Freund’s adjuvant (CFA) in sheep and goats. The animals were divided into four groups: CFA-treated sheep, control sheep, CFA-treated goat, and control goat groups (n 3 in each group). The dorsal root ganglions of these animals were isolated and used for the construction of a cDNA= library and transcriptome sequencing. Diferentially expressed genes (DEGs) were identifed in CFA-induced sheep and goats and gene ontology (GO) enrichment analysis was performed. Results: In total, 1748 and 2441 DEGs were identifed in CFA-treated goat and sheep, respectively. The DEGs identi- fed in CFA-treated goats, such as C-C motif chemokine ligand 27 (CCL27), glutamate receptor 2 (GRIA2), and sodium voltage-gated channel alpha subunit 3 (SCN3A), were mainly enriched in GO functions associated with N-methyl- D-aspartate (NMDA) receptor, infammatory response, and immune response. The DEGs identifed in CFA-treated sheep, such as gamma-aminobutyric acid (GABA)-related DEGs (gamma-aminobutyric acid type A receptor gamma 3 subunit [GABRG3], GABRB2, and GABRB1), SCN9A, and transient receptor potential cation channel subfamily V member 1 (TRPV1), were mainly enriched in GO functions related to neuroactive ligand-receptor interaction, NMDA receptor, and defense response. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Transcriptional and Post-Transcriptional Regulation of ATP-Binding Cassette Transporter Expression
Transcriptional and Post-transcriptional Regulation of ATP-binding Cassette Transporter Expression by Aparna Chhibber DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Pharmaceutical Sciences and Pbarmacogenomies in the Copyright 2014 by Aparna Chhibber ii Acknowledgements First and foremost, I would like to thank my advisor, Dr. Deanna Kroetz. More than just a research advisor, Deanna has clearly made it a priority to guide her students to become better scientists, and I am grateful for the countless hours she has spent editing papers, developing presentations, discussing research, and so much more. I would not have made it this far without her support and guidance. My thesis committee has provided valuable advice through the years. Dr. Nadav Ahituv in particular has been a source of support from my first year in the graduate program as my academic advisor, qualifying exam committee chair, and finally thesis committee member. Dr. Kathy Giacomini graciously stepped in as a member of my thesis committee in my 3rd year, and Dr. Steven Brenner provided valuable input as thesis committee member in my 2nd year. My labmates over the past five years have been incredible colleagues and friends. Dr. Svetlana Markova first welcomed me into the lab and taught me numerous laboratory techniques, and has always been willing to act as a sounding board. Michael Martin has been my partner-in-crime in the lab from the beginning, and has made my days in lab fly by. Dr. Yingmei Lui has made the lab run smoothly, and has always been willing to jump in to help me at a moment’s notice.