Identification of Novel Catalytic Features of Endo-Β-1,4-Glucanase Produced by Mulberry Longicorn Beetle Apriona Germari
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
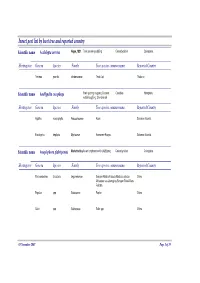
Insect Pest List by Host Tree and Reported Country
Insect pest list by host tree and reported country Scientific name Acalolepta cervina Hope, 1831 Teak canker grub|Eng Cerambycidae Coleoptera Hosting tree Genera Species Family Tree species common name Reported Country Tectona grandis Verbenaceae Teak-Jati Thailand Scientific name Amblypelta cocophaga Fruit spotting bug|eng Coconut Coreidae Hemiptera nutfall bug|Eng, Chinche del Hosting tree Genera Species Family Tree species common name Reported Country Agathis macrophylla Araucariaceae Kauri Solomon Islands Eucalyptus deglupta Myrtaceae Kamarere-Bagras Solomon Islands Scientific name Anoplophora glabripennis Motschulsky Asian longhorn beetle (ALB)|eng Cerambycidae Coleoptera Hosting tree Genera Species Family Tree species common name Reported Country Paraserianthes falcataria Leguminosae Sengon-Albizia-Falcata-Molucca albizia- China Moluccac sau-Jeungjing-Sengon-Batai-Mara- Falcata Populus spp. Salicaceae Poplar China Salix spp. Salicaceae Salix spp. China 05 November 2007 Page 1 of 35 Scientific name Aonidiella orientalis Newstead, Oriental scale|eng Diaspididae Homoptera 1894 Hosting tree Genera Species Family Tree species common name Reported Country Lovoa swynnertonii Meliaceae East African walnut Cameroon Azadirachta indica Meliaceae Melia indica-Neem Nigeria Scientific name Apethymus abdominalis Lepeletier, Tenthredinidae Hymenoptera 1823 Hosting tree Genera Species Family Tree species common name Reported Country Other Coniferous Other Coniferous Romania Scientific name Apriona germari Hope 1831 Long-horned beetle|eng Cerambycidae -

Ramesh Chander Bhagat.Pdf
Int. J. Curr. Res. Biosci. Plant Biol. 2016, 3(12): 115-123 International Journal of Current Research in Biosciences and Plant Biology ISSN: 2349-8080 (Online) ● Volume 3 ● Number 12 (December-2016) Journal homepage: www.ijcrbp.com Original Research Article doi: http://dx.doi.org/10.20546/ijcrbp.2016.312.014 An Update on Checklist and Biodiversity of Coleopteran-fauna (Insecta) of Forestry and Mulberry Importance in Jammu and Kashmir State (India) Ramesh Chander Bhagat* P.O. Box No. 1250, G.P.O., Residency Road, Srinagar, Kashmir-190 001, J & K, India *Corresponding author. A b s t r a c t Article Info The present paper deals with a total of 64 species of beetles and weevils (Coleoptera), Accepted: 29 November 2016 belonging to 52 genera, under 14 families, associated with diverse species of forest and Available Online: 06 December 2016 mulberry plantations, occurring in vast areas and localities of Jammu and Kashmir State. The Coleopteran species of forestry and mulberry importance accounts for 73.43 K e y w o r d s % and 35.93 % respectively. The Coleopteran-fauna (47 spp.), spread over 12 families, is found to be infesting forest trees,viz. Ash, Benne, Birch, Conifers, Elms, Biodiversity Ivy, Maple, Oak, Parrotia, Plane tree, Poplars, Robinia, Salix, and Yew. Of these trees, Checklist Pines showed highest number of Coleopteran species i.e. 18, under 6 families, followed Coleopteran-fauna by Poplars, with 15 spp. (4 families) and Cedars, having 14 spp. (4 families) The Forest trees Mulberry plantations Mulberry plantations (Morus spp.) both endemic as well as exotic, have been observed to be infesting 23 spp. -

PRA on Apriona Species
EUROPEAN AND MEDITERRANEAN PLANT PROTECTION ORGANIZATION ORGANISATION EUROPEENNE ET MEDITERRANEENNE POUR LA PROTECTION DES PLANTES 16-22171 (13-18692) Only the yellow note is new compared to document 13-18692 Pest Risk Analysis for Apriona germari, A. japonica, A. cinerea Note: This PRA started 2011; as a result, three species of Apriona were added to the EPPO A1 List: Apriona germari, A. japonica and A. cinerea. However recent taxonomic changes have occurred with significant consequences on their geographical distributions. A. rugicollis is no longer considered as a synonym of A. germari but as a distinct species. A. japonica, which was previously considered to be a distinct species, has been synonymized with A. rugicollis. Finally, A. cinerea remains a separate species. Most of the interceptions reported in the EU as A. germari are in fact A. rugicollis. The outcomes of the PRA for these pests do not change. However A. germari has a more limited and a more tropical distribution than originally assessed, but it is considered that it could establish in Southern EPPO countries. The Panel on Phytosanitary Measures agreed with the addition of Apriona rugicollis to the A1 list. September 2013 EPPO 21 Boulevard Richard Lenoir 75011 Paris www.eppo.int [email protected] This risk assessment follows the EPPO Standard PM PM 5/3(5) Decision-support scheme for quarantine pests (available at http://archives.eppo.int/EPPOStandards/pra.htm) and uses the terminology defined in ISPM 5 Glossary of Phytosanitary Terms (available at https://www.ippc.int/index.php). This document was first elaborated by an Expert Working Group and then reviewed by the Panel on Phytosanitary Measures and if relevant other EPPO bodies. -

Larval Peritrophic Matrix Yu-Bo Lin1,2†, Jing-Jing Rong1,2†, Xun-Fan Wei3, Zhuo-Xiao Sui3, Jinhua Xiao3 and Da-Wei Huang3*
Lin et al. Proteome Science (2021) 19:7 https://doi.org/10.1186/s12953-021-00175-x RESEARCH Open Access Proteomics and ultrastructural analysis of Hermetia illucens (Diptera: Stratiomyidae) larval peritrophic matrix Yu-Bo Lin1,2†, Jing-Jing Rong1,2†, Xun-Fan Wei3, Zhuo-Xiao Sui3, Jinhua Xiao3 and Da-Wei Huang3* Abstract Background: The black soldier fly (Hermetia illucens) has significant economic potential. The larvae can be used in financially viable waste management systems, as they are voracious feeders able to efficiently convert low-quality waste into valuable biomass. However, most studies on H. illucens in recent decades have focused on optimizing their breeding and bioconversion conditions, while information on their biology is limited. Methods: About 200 fifth instar well-fed larvae were sacrificed in this work. The liquid chromatography-tandem mass spectrometry and scanning electron microscopy were employed in this study to perform a proteomic and ultrastructural analysis of the peritrophic matrix (PM) of H. illucens larvae. Results: A total of 565 proteins were identified in the PM samples of H. illucen, of which 177 proteins were predicted to contain signal peptides, bioinformatics analysis and manual curation determined 88 proteins may be associated with the PM, with functions in digestion, immunity, PM modulation, and others. The ultrastructure of the H. illucens larval PM observed by scanning electron microscopy shows a unique diamond-shaped chitin grid texture. Conclusions: It is the first and most comprehensive proteomics research about the PM of H. illucens larvae to date. All the proteins identified in this work has been discussed in details, except several unnamed or uncharacterized proteins, which should not be ignored and need further study. -

New Records of Longhorn Beetles (Cerambycidae: Coleoptera) From
The Journal of Zoology Studies 2016; 3(1): 19-26 The Journal of Zoology Studies ISSN 2348-5914 New records of longhorn beetles (Cerambycidae: Coleoptera) JOZS 2016; 3(1): 19-26 from Manipur State India with Checklist JOZS © 2016 Received: 11-02-2016 Author: Bulganin Mitra, Priyanka Das, Kaushik Mallick, Udipta Chakraborti and Amitava Majumder Accepted: 22-03-2016 Abstract Bulganin Mitra, Present communication reports 50 species of 40 genera under 24 tribes belonging to 5 Scientist – C Zoological Survey of India, Subfamilies of the family Cerambycidae from the state of Manipur. Among them, Dorysthenes Kolkata – 700053. India (Lophosternus) huegelii (Redtenbacher, 1848) and Olenecamptus bilobus (Fabricius, 1801) are Priyanka Das recorded for the first time from Manipur state. Moreover, 20 % of the total cerambycid fauna is Junior Research Fellow, Zoological Survey of India, restricted/endemic to the state. Kolkata – 700053. India Keywords: India, Manipur, Cerambycidae, New record Kaushik Mallick Post Graduate Department of 1. Introduction Zoology, Ashutosh College, Manipur is one of the seven states of Northeast India, lies at a latitude of 23°83′ N to 25°68′ N Kolkata – 700026. and a longitude of 93°03′ E to 94°78′ E. The total area covered by the state is 22,347 km². The India natural vegetation occupies an area of about 14,365 km² which is nearly 64% of the total Udipta Chakraborti, geographical area of the state and which attracts a large number of forest pests. Junior Research Fellow, Zoological Survey of India, Kolkata – 700053. The members of the family Cerambycidae are commonly known as longhorn beetles or round- India headed borer beetles, is one of the notorious groups of insect pest due to their diurnal and Amitava Majumder nocturnal activities. -

Variation in Inspection Efficacy by Member States of Wood Packaging Material Entering the European Union
Journal of Economic Entomology, 111(2), 2018, 707–715 doi: 10.1093/jee/tox357 Advance Access Publication Date: 20 January 2018 Forest Entomology Research Article Variation in Inspection Efficacy by Member States of Wood Packaging Material Entering the European Union Dominic Eyre,1,5 Roy Macarthur,2 Robert A. Haack,3 Yi Lu,2 and Hannes Krehan4 1Defra, Sand Hutton, York YO41 1LZ, UK, 2Fera Science Ltd., Sand Hutton, York YO41 1LZ, UK, 3USDA Forest Service, Northern Research Station, 3101 Technology Boulevard, Suite F, Lansing, MI 48910 (Emeritus), 4Bundesamt für Wald, Seckendorff- Gudent-Weg 8, 1131 Wien, Austria, and 5Corresponding author, e-mail: [email protected] Subject Editor: Lisa Neven Received 31 August 2017; Editorial decision 21 November 2017 Abstract The use of wood packaging materials (WPMs) in international trade is recognized as a pathway for the movement of invasive pests and as the origin of most introductions of Asian longhorned beetle, Anoplophora glabripennis (Motschulsky) (Coleoptera: Cerambycidae) in Europe and North America. Following several pest interceptions on WPM associated with stone imports from China, the European Union (EU) agreed to survey certain categories of imports based on the EU Combined Nomenclature Codes for imports, which are based on the international Harmonized System. Between April 2013 and March 2015, 72,263 relevant consignments were received from China in the EU and 26,008 were inspected. Harmful organisms were detected in 0.9% of the consignments, and 1.1% of the imports did not have markings compliant with the international standard for treating WPM, ISPM 15. There were significant differences between the detection rates of harmful organisms among EU member states. -

The Major Arthropod Pests and Weeds of Agriculture in Southeast Asia
The Major Arthropod Pests and Weeds of Agriculture in Southeast Asia: Distribution, Importance and Origin D.F. Waterhouse (ACIAR Consultant in Plant Protection) ACIAR (Australian Centre for International Agricultural Research) Canberra AUSTRALIA The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. Its mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Australian and developing country researchers in fields where Australia has a special research competence. Where trade names are used this constitutes neither endorsement of nor discrimination against any product by the Centre. ACIAR MO'lOGRAPH SERIES This peer-reviewed series contains the results of original research supported by ACIAR, or deemed relevant to ACIAR's research objectives. The series is distributed internationally, with an emphasis on the Third World. © Australian Centre for 1I1lernational Agricultural Resl GPO Box 1571, Canberra, ACT, 2601 Waterhouse, D.F. 1993. The Major Arthropod Pests an Importance and Origin. Monograph No. 21, vi + 141pI- ISBN 1 86320077 0 Typeset by: Ms A. Ankers Publication Services Unit CSIRO Division of Entomology Canberra ACT Printed by Brown Prior Anderson, 5 Evans Street, Burwood, Victoria 3125 ii Contents Foreword v 1. Abstract 2. Introduction 3 3. Contributors 5 4. Results 9 Tables 1. Major arthropod pests in Southeast Asia 10 2. The distribution and importance of major arthropod pests in Southeast Asia 27 3. The distribution and importance of the most important arthropod pests in Southeast Asia 40 4. Aggregated ratings for the most important arthropod pests 45 5. Origin of the arthropod pests scoring 5 + (or more) or, at least +++ in one country or ++ in two countries 49 6. -

Edible Insects
1.04cm spine for 208pg on 90g eco paper ISSN 0258-6150 FAO 171 FORESTRY 171 PAPER FAO FORESTRY PAPER 171 Edible insects Edible insects Future prospects for food and feed security Future prospects for food and feed security Edible insects have always been a part of human diets, but in some societies there remains a degree of disdain Edible insects: future prospects for food and feed security and disgust for their consumption. Although the majority of consumed insects are gathered in forest habitats, mass-rearing systems are being developed in many countries. Insects offer a significant opportunity to merge traditional knowledge and modern science to improve human food security worldwide. This publication describes the contribution of insects to food security and examines future prospects for raising insects at a commercial scale to improve food and feed production, diversify diets, and support livelihoods in both developing and developed countries. It shows the many traditional and potential new uses of insects for direct human consumption and the opportunities for and constraints to farming them for food and feed. It examines the body of research on issues such as insect nutrition and food safety, the use of insects as animal feed, and the processing and preservation of insects and their products. It highlights the need to develop a regulatory framework to govern the use of insects for food security. And it presents case studies and examples from around the world. Edible insects are a promising alternative to the conventional production of meat, either for direct human consumption or for indirect use as feedstock. -

Research Article Morphological and Molecular Observation to Confirm
Jurnal Perlindungan Tanaman Indonesia, Vol. 21, No. 2, 2017: 96–105 DOI: 10.22146/jpti.25748 Research Article Morphological and Molecular Observation to Confirm the Taxonomic of Coptocercus biguttatus (Coleoptera: Cerambycidae) on Cloves in Ambon and Part of Ceram Island Observasi Morfologi dan Molekuler untuk Konfirmasi Taksonomi Coptocercus biguttatus (Coleoptera: Cerambycidae) pada Tanaman Cengkih di Pulau Ambon dan Sebagian Pulau Seram Mohamad Pamuji Setyolaksono1)*, Suputa2), & Nugroho Susetya Putra 2) 1)Regional Estate Crop Office on Seed Management and Plant Protection, Ambon Jln. Pertanian-Passo, Ambon 97232 2)Department of Crop Protection, Faculty of Agriculture, Universitas Gadjah Mada, Jln. Flora 1, Bulaksumur, Sleman, Yogyakarta 55281 *Corresponding author. E-mail: [email protected] Submitted June 7, 2017; accepted August 9, 2017 ABSTRACT This research was conducted to confirm the species of longhorn beetle (Coptocercus biguttatus) drilling clove stems in Ambon and part of Ceram Island, Moluccas, which has been noted as an important pest. Aim of this investigation was to characterize the species morphologically, and more detailed with molecular technique via mtCO1 gene analysis. The longhorn beetle was taken in Ambon and part of Ceram Island, Moluccas and then was etablished in laboratory with host rearing method on pieces of clove stem. The results showed that C. biguttatus attacking clove stems in Ambon and part of Ceram Island, Maluku was closely related and grouped into same cluster with C. rubripes and P. semipunctata in of New Zealand with 85% homology value. C. biguttatus distributed evenly in all clove planting areas in Ambon and part of Ceram Island. Keywords: Cerambycidae, clove stem borer, Coptocercus biguttatus, Mollucas INTISARI Penelitian ini dilakukan untuk mengonfirmasi spesies kumbang sungut panjang (Coptocercus biguttatus) yang menggerek batang tanaman cengkih di Pulau Ambon dan sebagian dari Pulau Seram, Maluku menggunakan karakter morfologi dan molekuler berbasis gen mtCO1 untuk sidik kekerabatan. -

Final Report
Final Report Project Title (Acronym) Further Development of Risk Management for the EC listed Anoplophora species, A. chinensis and A. glabripennis (ANOPLORISK-II) Project Duration: Start date: 01/01/14 End date: 31/12/15 1. Research Consortium Partners Coordinator – Partner 1 Federal Research and Training Centre for Forests, Natural Hazards and Landscape Organisation (BFW); Department of Forest Protection Name of Contact Dr. Gernot Hoch Gender: M (incl. Title) Job Title Head of Department of Forest Protection Postal Address Seckendorff-Gudent-Weg 8, 1131 Vienna, Austria E-mail [email protected] Phone +43 1 87838-1155 Partner 2 Julius Kühn-Institut, Federal Research Centre for Cultivated Plants, Institute for Organisation National and International Plant Health (JKI) Name of Contact Dr. Thomas Schröder Gender: M (incl. Title) Job Title Senior Scientist Postal Address Messeweg 11/12, D.-38104 Braunschweig, Germany E-mail [email protected] Phone +49 531 299 3381 Partner 3 Organisation Fera Science Ltd Name of Contact Dr. Robert Weaver Gender: M (incl. Title) Job Title Head of Invertebrate Pest Management Postal Address Fera, Sand Hutton, York, YO41 1LZ, UK E-mail [email protected] Phone +44 1904 462627 ANOPLORISK-II Page 2 of 56 2. Executive Summary Project Summary Further Development of Risk Management for the EC listed Anoplophora species, A. chinensis and A. glabripennis (ANOPLORISK-II) There is a high threat of harmful impact arising from material infested by Anoplophora chinensis (Citrus longhorned beetle; CLB) and Anoplophora glabripennis (Asian longhorned beetle; ALB) within EU territory. While interceptions of CLB with plants for plantings have been reduced in the last years due to recommended measures according to the implementing decision 2012/138/EU of the EU Commission, ALB is intercepted in wood packaging material in continuously high numbers. -

The Checklist of Longhorn Beetles (Coleoptera: Cerambycidae) from India
Zootaxa 4345 (1): 001–317 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Monograph ZOOTAXA Copyright © 2017 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4345.1.1 http://zoobank.org/urn:lsid:zoobank.org:pub:1D070D1A-4F99-4EEF-BE30-7A88430F8AA7 ZOOTAXA 4345 The checklist of longhorn beetles (Coleoptera: Cerambycidae) from India B. KARIYANNA1,4, M. MOHAN2,5, RAJEEV GUPTA1 & FRANCESCO VITALI3 1Indira Gandhi Krishi Vishwavidyalaya, Raipur, Chhattisgarh-492012, India . E-mail: [email protected] 2ICAR-National Bureau of Agricultural Insect Resources, Bangalore, Karnataka-560024, India 3National Museum of Natural History of Luxembourg, Münster Rd. 24, L-2160 Luxembourg, Luxembourg 4Current address: University of Agriculture Science, Raichur, Karnataka-584101, India 5Corresponding author. E-mail: [email protected] Magnolia Press Auckland, New Zealand Accepted by Q. Wang: 22 Jun. 2017; published: 9 Nov. 2017 B. KARIYANNA, M. MOHAN, RAJEEV GUPTA & FRANCESCO VITALI The checklist of longhorn beetles (Coleoptera: Cerambycidae) from India (Zootaxa 4345) 317 pp.; 30 cm. 9 Nov. 2017 ISBN 978-1-77670-258-9 (paperback) ISBN 978-1-77670-259-6 (Online edition) FIRST PUBLISHED IN 2017 BY Magnolia Press P.O. Box 41-383 Auckland 1346 New Zealand e-mail: [email protected] http://www.mapress.com/j/zt © 2017 Magnolia Press All rights reserved. No part of this publication may be reproduced, stored, transmitted or disseminated, in any form, or by any means, without prior written permission from the publisher, to whom all requests to reproduce copyright material should be directed in writing. This authorization does not extend to any other kind of copying, by any means, in any form, and for any purpose other than private research use. -

Longhorn Beetles (Cerambycidae: Coleoptera) of Himachal Pradesh 405 ISSN 0375-1511
MITRA et al. : Longhorn beetles (Cerambycidae: Coleoptera) of Himachal Pradesh 405 ISSN 0375-1511 Rec. zool. Surv. India : 115(Part-4) : 405-409, 2015 LONGHORN BEETLES (CERAMBYCIDAE: COLEOPTERA) OF HIMACHAL PRADESH BULGANIN MITRA#, AMITAVA MAJUMDER, UDIPTA CHAKRABORTI, PRIYANKA DAS AND KAUSHIK MALLICK* Zoological Survey of India, Prani Vigyan Bhavan, M-Block, New Alipore, Kolkata-700 053, West Bengal, India. * Post Graduate Department of Zoology, Asutosh College, Kolkata – 700026 #Corresponding author. E-mail: [email protected] INTRODUCTION complete bibliography on the works of cerambycid The taxonomic study of the family beetles of this Himalayan state in India. cerambycidae is very poor in Himachal Pradesh. Subfamily LAMIINAE Contributions of Breuning (1937, 1958, 1965), Tribe ANCYLONOTINI Lacordaire, 1869 Beeson and Bhatia (1939), Basak and Biswas 1. Palimnodes ducalis (Bates, 1884) (1993), Mukhopadhyay (2011), Saha et al. (2013) 1884. Apalimna ducalis Bates, The Journal of the Linnean were enriched the cerambycid fauna of this state. Society of London. Zoology, 18: 242. Type Locality: Later, few publications were made on other North India, Type Repository: Muséum National aspects than taxonomy of cerambycidae by Uniyal d’Histoire Naturelle, Paris. and Mathur (1998), Singh, and Verma (1998), 2006. Parapalimna ducalis; Weigel, Biodiversität und Naturausstattung im Himalaya, II. V: 503. Bhargava and Uniyal (2011). Therefore, an attempt Material examined: 1 ex, Kulu, Himachal has been taken to prepare a consolidated taxonomic Pradesh,