Phenotypic, Physiological, and Genetic Patterns of TTX Resistance in the Sierra Garter Snake Thamnophis Couchii
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

State of Sierra Frogs
State of Sierra Frogs A report on the status of frogs & toads in the Sierra Nevada & California Cascade Mountains State of Sierra Frogs A report on the status of frogs & toads in the Sierra Nevada & California Cascade Mountains By Marion Gee, Sara Stansfield, & Joan Clayburgh July 2008 www.sierranevadaalliance.org State of Sierra Frogs 1 Acknowledgements The impetus for this report was the invaluable research on pesticides by Carlos Davidson, professor at San Francisco State University. Davidson, along with Amy Lind (US Forest Service), Curtis Milliron (California Department of Fish and Game), David Bradford (United States Environmental Protection Agency) and Kim Vincent (Graduate Student, San Francisco State University), generously donated their time and expertise to speak at two public workshops on the topics of Sierra frogs and toads as well as to provide comments for this document. Our thanks to the other reviewers of this manuscripts including Bob Stack (Jumping Frog Research Institute), Katie Buelterman, Dan Keenan, and Genevieve Jessop Marsh. This project was fortunate to receive contributions of photography and artwork from John Muir Laws, Elena DeLacy, Bob Stack, Ralph & Lisa Cutter and Vance Vredenburg. Photo credits are found with each caption. This work was made possible by generous grants from the Rose Foundation for Communities and the Environment and the State Water Resources Control Board. Funding for this project has been provided in part through an Agreement with the State Water Resources Control Board (SWRCB) pursuant to the Costa-Machado Water Act of 2000 (Proposition 13) and any amendments thereto for the implementation of California’s Non-point Source Pollution Control Program. -

What Are Their Roles in Mitochondrial Protein Synthesis?
Characterisation of human mtRF1 and C12orf65: What are their roles in mitochondrial protein synthesis? Aleksandra Pajak M.Res Thesis submitted to Newcastle University in candidature for the degree of Doctor of Philosophy Newcastle University Faculty of Medical Sciences Institute for Ageing and Health Mitochondrial Research Group January 2013 Abstract Mitochondria have their own protein synthesis machinery that synthesises the oxidative phosphorylation components encoded by their mtDNA. This translation process consists of four main phases: initiation, elongation, termination and ribosome recycling. Termination and its control have been the least investigated. Recently, however, the termination factor, mtRF1a, has been characterised as sufficient to release all the nascent proteins from the mitoribosome. Furthermore, bioinformatics has identified three additional members of this mitochondrial release factor family namely, mtRF1, C12orf65 and ICT1. The latter is now known to be incorporated into the mitoribosome but its exact function remains unclear. My project has therefore focussed on characterising the remaining two factors; mtRF1 and C12orf65, and investigating their possible involvement in mitochondrial protein synthesis. It has been demonstrated that protein synthesis is not perfect and bacterial ribosomes not infrequently stall during translation. This can result from limiting amounts of charged tRNAs, stable secondary structures, or truncated/degraded transcripts. Ribosome stalling has been shown to cause growth arrest. In order to prevent that and maintain high efficiency of mitochondrial protein synthesis such stalled complexes need to be rapidly recycled. Bacteria have developed at least three distinct mechanisms by which ribosomes can be rescued. Contrastingly, despite the presence of truncated mRNAs in mitochondria, no such quality control mechanisms have been identified in these organelles. -

Mountain Yellow-Legged Frog (Rana Muscosa and Rana Sierrae) As Endangered Under the California Endangered Species Act
BEFORE THE CALIFORNIA FISH AND GAME COMMISSION A Petition to List All Populations of the Mountain Yellow-Legged Frog (Rana muscosa and Rana sierrae) as Endangered under the California Endangered Species Act Photo © Todd Vogel CENTER FOR BIOLOGICAL DIVERSITY, PETITIONER January 25, 2010 Petition to California Fish & Game Commission to List the Mountain Yellow-Legged Frog as Endangered Center for Biological Diversity January 25, 2010 Notice of Petition For action pursuant to Section 670.1, Title 14, California Code of Regulations (CCR) and Sections 2072 and 2073 of the Fish and Game Code relating to listing and delisting endangered and threatened species of plants and animals. I. SPECIES BEING PETITIONED: Common Name: mountain yellow-legged frog (southern mountain yellow-legged frog and Sierra Nevada mountain yellow-legged frog) Scientific Name: Rana muscosa and Rana sierrae II. RECOMMENDED ACTION: List as Endangered The Center for Biological Diversity submits this petition to list all populations of the mountain yellow-legged frog in California the as endangered throughout their range in California, under the California Endangered Species Act (California Fish and Game Code §§ 2050 et seq., “CESA”). This petition demonstrates that the both the southern mountain yellow-legged frog (Rana muscosa) and the Sierra Nevada mountain yellow-legged frog (Rana sierrae) clearly warrant listing under CESA based on the factors specified in the statute. III. AUTHOR OF PETITION: Name: Lisa Belenky, Senior Attorney, Center For Biological Diversity (with the assistance of Ellen Howard, B.A. EPO Biology, University of Colorado) Address: 351 California Street, Suite 600, San Francisco, CA 94104 Phone: 415-436-9682 x 307 Fax: 415-436-9683 Email: [email protected] I hereby certify that, to the best of my knowledge, all statements made in this petition are true and complete. -

Analysis of the Relationship Between Ribosomal Protein and SSU Processome Assembly in Saccharomyces Cerevisiae
Analysis of the relationship between ribosomal protein and SSU processome assembly in Saccharomyces cerevisiae Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der naturwissenschaftlichen Fakultät III – Biologie und vorklinische Medizin - der Universität Regensburg vorgelegt von Steffen Jakob aus Wolfen Januar 2010 Promotionsgesuch eingereicht am: 13. Januar 2010 Die Arbeit wurde angeleitet von: Prof. Dr. Herbert Tschochner Prüfungsausschuss: Vorsitzender: Prof. Dr. Armin Kurtz 1. Prüfer: Prof. Dr. Herbert Tschochner 2. Prüfer: Prof. Dr. Rainer Deutzmann 3. Prüfer: Prof. Dr. Wolfgang Seufert Tag der mündlichen Prüfung: 24. März 2010 Die vorliegende Arbeit wurde in der Zeit von April 2006 bis Januar 2010 am Lehrstuhl Biochemie III des Institutes für Biochemie, Genetik und Mikrobiologie der Naturwissenschaftlichen Fakultät III der Universität zu Regensburg unter Anleitung von Dr. Philipp Milkereit im Labor von Prof. Dr. Herbert Tschochner angefertigt. Ich erkläre hiermit, dass ich diese Arbeit selbst verfasst und keine anderen als die angegebenen Quellen und Hilfsmittel verwendet habe. Diese Arbeit war bisher noch nicht Bestandteil eines Prüfungsverfahrens. Andere Promotionsversuche wurden nicht unternommen. Regensburg, den 13. Januar 2010 Steffen Jakob Table of Contents Table of Contents 1 SUMMARY ...................................................................................................... 1 2 INTRODUCTION ............................................................................................ -
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
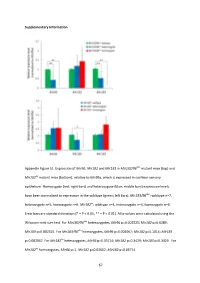
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Micrornas Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins
cells Review microRNAs Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins Abu Musa Md Talimur Reza 1,2 and Yu-Guo Yuan 1,3,* 1 Jiangsu Co-Innovation Center of Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou 225009, China; [email protected] 2 Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawi´nskiego5a, 02-106 Warsaw, Poland 3 Jiangsu Key Laboratory of Zoonosis/Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou 225009, China * Correspondence: [email protected]; Tel.: +86-514-8797-9228 Abstract: Ribosomal proteins (RPs) are mostly derived from the energy-consuming enzyme families such as ATP-dependent RNA helicases, AAA-ATPases, GTPases and kinases, and are important structural components of the ribosome, which is a supramolecular ribonucleoprotein complex, composed of Ribosomal RNA (rRNA) and RPs, coordinates the translation and synthesis of proteins with the help of transfer RNA (tRNA) and other factors. Not all RPs are indispensable; in other words, the ribosome could be functional and could continue the translation of proteins instead of lacking in some of the RPs. However, the lack of many RPs could result in severe defects in the biogenesis of ribosomes, which could directly influence the overall translation processes and global expression of the proteins leading to the emergence of different diseases including cancer. While microRNAs (miRNAs) are small non-coding RNAs and one of the potent regulators of the post-transcriptional 0 gene expression, miRNAs regulate gene expression by targeting the 3 untranslated region and/or coding region of the messenger RNAs (mRNAs), and by interacting with the 50 untranslated region, Citation: Reza, A.M.M.T.; Yuan, Y.-G. -

Target Species Mapping for the Green Visions Plan
Target Species Habitat Mapping California Quail and Mountain Quail (Callipepla californica and Oreortyx pictus) Family: Phasianidae Order: Galliformes Class: Aves WHR #: B140 and B141 Distribution: California quail are found in southern Oregon, northern Nevada, California, and Baja California, and have been introduced in other states such as Hawaii, Washington, Idaho, Colorado, and Utah (Peterson 1961). In California, they are widespread but absent from the higher elevations of the Sierra Nevada, the Cascades, the White Mountains, and the Warner Mountains, and are replaced by the related Gambel’s quail (C. gambelii) in some desert regions (Peterson 1961, Small 1994). In southern California, they are found from the Coast Range south to the Mexican border, and occur as far east as the western fringes of the Mojave and Sonoran deserts, such as in the Antelope Valley (Garrett and Dunn 1981, Small 1994). California quail range from sea level to about 5000 ft (1524 meters; Stephenson and Calcarone 1999) Mountain quail are resident from northern Washington and northern Idaho, south through parts of Oregon, northwestern Nevada, California, and northern Baja California (Peterson 1961). In southern California, mountain quail are found in nearly all of the mountain ranges west of the deserts, including the southern Coast Ranges, from the Santa Lucia Mountains south through Santa Barbara and Ventura counties, and the Peninsular Ranges south to the Mexican border (Garrot and Dunn 1981, Small 1994). In the Transverse Ranges, a small population occurs in the western Santa Monica Mountains, and larger populations occur in the San Gabriel and San Bernardino Mountains (Small 1994). Mountain quail are found at elevations from below 2000 ft (610 meters) to over 9000 ft (2743 meters; Stephenson and Calcarone 1999). -

History of the Ribosome and the Origin of Translation
History of the ribosome and the origin of translation Anton S. Petrova,1, Burak Gulena, Ashlyn M. Norrisa, Nicholas A. Kovacsa, Chad R. Berniera, Kathryn A. Laniera, George E. Foxb, Stephen C. Harveyc, Roger M. Wartellc, Nicholas V. Huda, and Loren Dean Williamsa,1 aSchool of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332; bDepartment of Biology and Biochemistry, University of Houston, Houston, TX, 77204; and cSchool of Biology, Georgia Institute of Technology, Atlanta, GA 30332 Edited by David M. Hillis, The University of Texas at Austin, Austin, TX, and approved November 6, 2015 (received for review May 18, 2015) We present a molecular-level model for the origin and evolution of building up of the functional centers, proceeds to the establishment the translation system, using a 3D comparative method. In this model, of the common core, and continues to the development of large the ribosome evolved by accretion, recursively adding expansion metazoan rRNAs. segments, iteratively growing, subsuming, and freezing the rRNA. Incremental evolution of function is mapped out by stepwise Functions of expansion segments in the ancestral ribosome are accretion of rRNA. In the extant ribosome, specific segments of assigned by correspondence with their functions in the extant rRNA perform specific functions including peptidyl transfer, ribosome. The model explains the evolution of the large ribosomal subunit association, decoding, and energy-driven translocation subunit, the small ribosomal subunit, tRNA, and mRNA. Prokaryotic (11). The model assumes that the correlations of rRNA segments ribosomes evolved in six phases, sequentially acquiring capabilities with their functions have been reasonably maintained over the for RNA folding, catalysis, subunit association, correlated evolution, broad course of ribosomal evolution. -

HUMAN RIBOSOME BIOGENESIS and the REGULATION of the TUMOUR SUPPRESSOR P53
HUMAN RIBOSOME BIOGENESIS AND THE REGULATION OF THE TUMOUR SUPPRESSOR p53 Andria Pelava Submitted for Doctor of Philosophy Final submission: December 2016 Institute of Cell and Molecular Biosciences Faculty of Medical Sciences Newcastle University ii Abstract Ribosome production is an energetically expensive and, therefore, highly regulated process. Defects in ribosome biogenesis lead to genetic diseases called Ribosomopathies, such as Dyskeratosis Congenita (DC), and mutations in ribosomal proteins and ribosome biogenesis factors are linked to multiple types of cancer. During ribosome biogenesis, the ribosomal RNAs (rRNAs) are processed and modified, and defects in ribosome biogenesis lead to the activation of p53. This project aimed to investigate the functions of Dyskerin, mutated in X-linked DC, in human ribosome biogenesis and p53 regulation, and to explore the link between ribosome production and p53 homeostasis. Dyskerin is an rRNA pseudouridine synthase and required for telomere maintenance. There is some debate as to whether DC is the result of telomere maintenance or ribosome biogenesis defects. It is shown here that human Dyskerin is required for the production of both LSU and SSU, and knockdown of Dyskerin leads to p53 activation via inhibition of MDM2 via the 5S RNP, an LSU assembly intermediate which accumulates after ribosome biogenesis defects. My data indicate that p53 activation, due to defects in ribosome biogenesis, may contribute to the clinical symptoms seen in patients suffering with DC. In addition, it is shown that defects in early or late stages of SSU or LSU biogenesis, result in activation of p53 via the 5S RNP-MDM2 pathway, and that p53 is induced in less than 12 hours after ribosome biogenesis defects. -

Here Describe This Pattern of Absence and Lay out a Plan for Restoring the Species to Areas Where the Species Has Recently Gone Extinct
San Joaquin Valley Natural Communities Conference March 25, 2021 Virtual Meeting via WHOVA Video Conferencing Program and Abstracts San Joaquin Valley Natural Communities Conference March 25, 2021 8:30 - 8:50 Brian Cypher Welcome to the Conference, Moderator 8:50 - 9:10 Xerónimo Castañeda Six years of Tricolored Blackbird Conservation in the San Joaquin Valley: Challenges and opportunities to save this imperiled species 9:10 - 9:30 Petros Chrysafis Scent deterrent as an applicable predator deterrent 9:30 -9:50 Mike Westphal Blunt-nosed Leopard Lizard occupancy in the Northern Clade: How do we reverse the recent pattern of extinction? 9:50 - 10:10 Deborah Woollett Guidelines, applications, and caveats to using detection dogs to find Blunt-nosed Leopard Lizard scat 10:10 – 10:30 BREAK 10:30 - 10:50 Erica Kelly Impact of a Sarcoptic Mange Epidemic on a Population of Endangered San Joaquin Kit Foxes 10:50 – 11:30 Nicole Deatherage Urban Landscape Attributes and Competition Affect San Joaquin Kit Fox Occupancy and Spatiotemporal Activity 11:30 – 12:00 LUNCH BREAK 12:00 – 12:30 Quick Talk Presentations Anna Doty Effects of fire on summer roost selection and torpor expression of bats in Sequoia and Kings Canyon National Parks: a planned project Patrick Walker Central Valley California tiger salamander behavior within vernal pool grassland complex in Madera County Christopher Lortie Too much of a good thing: richness and restoration in drylands Petros Chrysafis Contribution to eMammal’s Snapshot USA Project Kristie Stein Tricolored Blackbird -

Species List for Sierra Nevada Lakes ( Compiled by Roland Knapp - Version 01 November 2018
Species List for Sierra Nevada Lakes (http://mountainlakesresearch.com/lake-fauna/) Compiled by Roland Knapp - version 01 November 2018 VERTEBRATES Phylum Class Order Family Genus & Species Comments Chordata Amphibia Anura Bufonidae Bufo (Anaxyrus) boreas halophilus California Toad Chordata Amphibia Anura Bufonidae Bufo (Anaxyrus) canorus Yosemite Toad Chordata Amphibia Anura Hylidae Pseudacris (Hyliola) regilla Pacific Treefrog Chordata Amphibia Anura Ranidae Rana muscosa Southern Mountain Yellow-legged Frog Chordata Amphibia Anura Ranidae Rana sierrae Sierra Nevada Yellow-legged Frog Chordata Amphibia Caudata Salamandridae Hydromantes platycephalus Mount Lyell Salamander Chordata Amphibia Caudata Salamandridae Taricha sierrae Sierra Newt Chordata Amphibia Caudata Salamandridae Taricha torosa California Newt Chordata Mammalia Carnivora Mustelidae Lontra canadensis Northern River Otter Chordata Mammalia Soricomorpha Soricidae Sorex palustris Northern Water Shrew Chordata Reptilia Squamata Colubridae Thamnophis couchi Sierra Garter Snake Chordata Reptilia Squamata Colubridae Thamnophis elegans elegans Mountain Garter Snake Chordata Reptilia Squamata Colubridae Thamnophis sirtalis fitchi Valley Garter Snake Chordata Reptilia Testudines Emydidae Actinemys marmorata Pacific Pond Turtle BENTHIC MACROINVERTEBRATES Phylum Class Order Family Genus & Species Comments Annelida Clitellata Arhynchobdellida Erpobdellidae Erpobdella punctata Annelida Clitellata Arhynchobdellida Erpobdellidae Mooreobdella microstoma Annelida Clitellata Arhynchobdellida