FOS Expression in Osteoid Osteoma and Osteoblastoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Infiltrating Myeloid Cells Drive Osteosarcoma Progression Via GRM4 Regulation of IL23
Published OnlineFirst September 16, 2019; DOI: 10.1158/2159-8290.CD-19-0154 RESEARCH BRIEF Infi ltrating Myeloid Cells Drive Osteosarcoma Progression via GRM4 Regulation of IL23 Maya Kansara 1 , 2 , Kristian Thomson 1 , Puiyi Pang 1 , Aurelie Dutour 3 , Lisa Mirabello 4 , Francine Acher5 , Jean-Philippe Pin 6 , Elizabeth G. Demicco 7 , Juming Yan 8 , Michele W.L. Teng 8 , Mark J. Smyth 9 , and David M. Thomas 1 , 2 ABSTRACT The glutamate metabotropic receptor 4 (GRM4 ) locus is linked to susceptibility to human osteosarcoma, through unknown mechanisms. We show that Grm4 − / − gene– targeted mice demonstrate accelerated radiation-induced tumor development to an extent comparable with Rb1 +/ − mice. GRM4 is expressed in myeloid cells, selectively regulating expression of IL23 and the related cytokine IL12. Osteosarcoma-conditioned media induce myeloid cell Il23 expression in a GRM4-dependent fashion, while suppressing the related cytokine Il12 . Both human and mouse osteosarcomas express an increased IL23:IL12 ratio, whereas higher IL23 expression is associated with worse survival in humans. Con- sistent with an oncogenic role, Il23−/− mice are strikingly resistant to osteosarcoma development. Agonists of GRM4 or a neutralizing antibody to IL23 suppressed osteosarcoma growth in mice. These fi ndings identify a novel, druggable myeloid suppressor pathway linking GRM4 to the proinfl ammatory IL23/IL12 axis. SIGNIFICANCE: Few novel systemic therapies targeting osteosarcoma have emerged in the last four decades. Using insights gained from a genome-wide association study and mouse modeling, we show that GRM4 plays a role in driving osteosarcoma via a non–cell-autonomous mechanism regulating IL23, opening new avenues for therapeutic intervention. -

Clinical Features of Benign Tumors of the External Auditory Canal According to Pathology
Central Annals of Otolaryngology and Rhinology Research Article *Corresponding author Jae-Jun Song, Department of Otorhinolaryngology – Head and Neck Surgery, Korea University College of Clinical Features of Benign Medicine, 148 Gurodong-ro, Guro-gu, Seoul, 152-703, South Korea, Tel: 82-2-2626-3191; Fax: 82-2-868-0475; Tumors of the External Auditory Email: Submitted: 31 March 2017 Accepted: 20 April 2017 Canal According to Pathology Published: 21 April 2017 ISSN: 2379-948X Jeong-Rok Kim, HwibinIm, Sung Won Chae, and Jae-Jun Song* Copyright Department of Otorhinolaryngology-Head and Neck Surgery, Korea University College © 2017 Song et al. of Medicine, South Korea OPEN ACCESS Abstract Keywords Background and Objectives: Benign tumors of the external auditory canal (EAC) • External auditory canal are rare among head and neck tumors. The aim of this study was to analyze the clinical • Benign tumor features of patients who underwent surgery for an EAC mass confirmed as a benign • Surgical excision lesion. • Recurrence • Infection Methods: This retrospective study involved 53 patients with external auditory tumors who received surgical treatment at Korea University, Guro Hospital. Medical records and evaluations over a 10-year period were examined for clinical characteristics and pathologic diagnoses. Results: The most common pathologic diagnoses were nevus (40%), osteoma (13%), and cholesteatoma (13%). Among the five pathologic subgroups based on the origin organ of the tumor, the most prevalent pathologic subgroup was the skin lesion (47%), followed by the epithelial lesion (26%), and the bony lesion (13%). No significant differences were found in recurrence rate, recurrence duration, sex, or affected side between pathologic diagnoses. -

Bone and Soft Tissue Tumors Have Been Treated Separately
EPIDEMIOLOGY z Sarcomas are rare tumors compared to other BONE AND SOFT malignancies: 8,700 new sarcomas in 2001, with TISSUE TUMORS 4,400 deaths. z The incidence of sarcomas is around 3-4/100,000. z Slight male predominance (with some subtypes more common in women). z Majority of soft tissue tumors affect older adults, but important sub-groups occur predominantly or exclusively in children. z Incidence of benign soft tissue tumors not known, but Fabrizio Remotti MD probably outnumber malignant tumors 100:1. BONE AND SOFT TISSUE SOFT TISSUE TUMORS TUMORS z Traditionally bone and soft tissue tumors have been treated separately. z This separation will be maintained in the following presentation. z Soft tissue sarcomas will be treated first and the sarcomas of bone will follow. Nowhere in the picture….. DEFINITION Histological z Soft tissue pathology deals with tumors of the classification connective tissues. of soft tissue z The concept of soft tissue is understood broadly to tumors include non-osseous tumors of extremities, trunk wall, retroperitoneum and mediastinum, and head & neck. z Excluded (with a few exceptions) are organ specific tumors. 1 Histological ETIOLOGY classification of soft tissue tumors tumors z Oncogenic viruses introduce new genomic material in the cell, which encode for oncogenic proteins that disrupt the regulation of cellular proliferation. z Two DNA viruses have been linked to soft tissue sarcomas: – Human herpes virus 8 (HHV8) linked to Kaposi’s sarcoma – Epstein-Barr virus (EBV) linked to subtypes of leiomyosarcoma z In both instances the connection between viral infection and sarcoma is more common in immunosuppressed hosts. -

CASE REPORT Intradermal Nevus of the External Auditory Canal
Int. Adv. Otol. 2009; 5:(3) 401-403 CASE REPORT Intradermal Nevus of the External Auditory Canal: A Case Report Sedat Ozturkcan, Ali Ekber, Riza Dundar, Filiz Gulustan, Demet Etit, Huseyin Katilmis Department of Otorhinolaryngology and Head and Neck Surgery ‹zmir Atatürk Research and Training Hospital, Ministry of Health, ‹ZM‹R-TURKEY (SO, AE, FG, DE, HK) Department of Otorhinolaryngology and Head and Neck Surgery Etimesgut Military Hospital , ANKARA-TURKEY (RD) Intradermal nevus is the most common skin tumor in humans; however, its occurrence in the external auditory canal (EAC) is uncommon. The clinical manifestations of pigmented nevus of the EAC have been reported to include ear fullness, foreign body sensation, hearing impairment, and otalgia, but some cases were asymptomatic and were found incidentally. The treatment of choice for a symptomatic intradermal nevus in the EAC is complete excision. There has been no recurrence reported in the literature . A pedunculated, papillomatous hair-bearing lesion was detected in the external auditory canal of the patient who was on follow-up for pruritus. Clinical and pathologic features of an intradermal nevus of the external auditory canal are presented, and the literature reviewed. Submitted : 14 October 2008 Revised : 01 July 2009 Accepted : 09 July 2009 Intradermal nevus is the most common skin tumor in left external auditory canal. Otomicroscopic humans; however, its occurrence in the external examination revealed a pedunculated, papillomatous auditory canal (EAC) is uncommon [1-4]. Intradermal hair-bearing lesion in the postero-inferior cartilaginous nevus is considered to be a form of benign cutaneous portion of the external auditory canal (Figure 1). -

Primary Ewing Sarcoma/Primitive Neuroectodermal Tumor of the Kidney: the MD Anderson Cancer Center Experience
cancers Article Primary Ewing Sarcoma/Primitive Neuroectodermal Tumor of the Kidney: The MD Anderson Cancer Center Experience Nidale Tarek 1,2,*, Rabih Said 3, Clark R. Andersen 4, Tina S. Suki 1, Jessica Foglesong 1,5 , Cynthia E. Herzog 1, Nizar M. Tannir 6, Shreyaskumar Patel 7, Ravin Ratan 7, Joseph A. Ludwig 7 and Najat C. Daw 1,* 1 Department of Pediatrics, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA; [email protected] (T.S.S.); [email protected] (J.F.); [email protected] (C.E.H.) 2 Department of Pediatrics and Adolescent Medicine, American University of Beirut Medical Center, Beirut 1107, Lebanon 3 Department of Investigational Cancer Therapeutics, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA; [email protected] 4 Department of Biostatistics, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA; [email protected] 5 Division of Hematology, Oncology, Neuro-Oncology & Stem Cell Transplant, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL 60611, USA 6 Department of Genitourinary Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA; [email protected] 7 Department of Sarcoma Medical Oncology, the University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA; [email protected] (S.P.); [email protected] (R.R.); [email protected] (J.A.L.) * Correspondence: [email protected] (N.T.); [email protected] (N.C.D.); Tel.: +1-713-792-6620 (N.C.D.) Received: 27 August 2020; Accepted: 5 October 2020; Published: 11 October 2020 Simple Summary: The Ewing sarcoma family of tumors (ESFT)s rarely originate in the kidneys and their treatment is significantly different from the other common kidney tumors. -
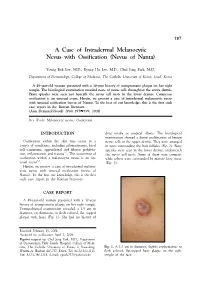
A Case of Intradermal Melanocytic Nevus with Ossification (Nevus of Nanta)
197 A Case of Intradermal Melanocytic Nevus with Ossification (Nevus of Nanta) Young Bok Lee, M.D., Kyung Ho Lee, M.D., Chul Jong Park, M.D. Department of Dermatology, College of Medicine, The Catholic University of Korea, Seoul, Korea A 49-year-old woman presented with a 30-year history of asymptomatic plaque on her right temple. The histological examination revealed nests of nevus cells throughout the entire dermis. Bony spicules were seen just beneath the nevus cell nests in the lower dermis. Cutaneous ossification is an unusual event. Herein, we present a case of intradermal melanocytic nevus with unusual ossification (nevus of Nanta). To the best of our knowledge, this is the first such case report in the Korean literature. (Ann Dermatol (Seoul) 20(4) 197∼199, 2008) Key Words: Melanocytic nevus, Ossification INTRODUCTION drug intake or medical illness. The histological examination showed a dense proliferation of benign Ossification within the skin may occur in a nevus cells in the upper dermis. They were arranged variety of conditions, including pilomatricoma, basal in nests surrounding the hair follicles (Fig. 2). Bony cell carcinoma, appendageal and fibrous prolifera- spicules were seen in the lower dermis, underneath 1,2 tion, inflammation and trauma . The occurrence of the nevus cell nests. Some of them were compact ossification within a melanocytic nevus is an un- while others were surrounded by mature fatty tissue 3-5 usual event . (Fig. 3). Herein, we present a case of intradermal melano- cytic nevus with unusual ossification (nevus of Nanta). To the best our knowledge, this is the first such case report in the Korean literature. -

Pathogenesis and Current Treatment of Osteosarcoma: Perspectives for Future Therapies
Journal of Clinical Medicine Review Pathogenesis and Current Treatment of Osteosarcoma: Perspectives for Future Therapies Richa Rathore 1 and Brian A. Van Tine 1,2,3,* 1 Division of Medical Oncology, Washington University in St. Louis, St. Louis, MO 63110, USA; [email protected] 2 Division of Pediatric Hematology and Oncology, St. Louis Children’s Hospital, St. Louis, MO 63110, USA 3 Siteman Cancer Center, St. Louis, MO 63110, USA * Correspondence: [email protected] Abstract: Osteosarcoma is the most common primary malignant bone tumor in children and young adults. The standard-of-care curative treatment for osteosarcoma utilizes doxorubicin, cisplatin, and high-dose methotrexate, a standard that has not changed in more than 40 years. The development of patient-specific therapies requires an in-depth understanding of the unique genetics and biology of the tumor. Here, we discuss the role of normal bone biology in osteosarcomagenesis, highlighting the factors that drive normal osteoblast production, as well as abnormal osteosarcoma development. We then describe the pathology and current standard of care of osteosarcoma. Given the complex hetero- geneity of osteosarcoma tumors, we explore the development of novel therapeutics for osteosarcoma that encompass a series of molecular targets. This analysis of pathogenic mechanisms will shed light on promising avenues for future therapeutic research in osteosarcoma. Keywords: osteosarcoma; mesenchymal stem cell; osteoblast; sarcoma; methotrexate Citation: Rathore, R.; Van Tine, B.A. Pathogenesis and Current Treatment of Osteosarcoma: Perspectives for Future Therapies. J. Clin. Med. 2021, 1. Introduction 10, 1182. https://doi.org/10.3390/ Osteosarcomas are the most common pediatric and adult bone tumor, with more than jcm10061182 1000 new cases every year in the United States alone. -

Osteoid Osteoma: Contemporary Management
eCommons@AKU Section of Orthopaedic Surgery Department of Surgery 2018 Osteoid osteoma: Contemporary management Shahryar Noordin Aga Khan University, [email protected] Salim Allana Emory University Kiran Hilal Aga Khan University, [email protected] Riaz Hussain Lukhadwala Aga Khan University, [email protected] Anum Sadruddin Pidani Aga Khan University, [email protected] See next page for additional authors Follow this and additional works at: https://ecommons.aku.edu/pakistan_fhs_mc_surg_orthop Part of the Orthopedics Commons, Radiology Commons, and the Surgery Commons Recommended Citation Noordin, S., Allana, S., Hilal, K., Lukhadwala, R. H., Pidani, A. S., Ud Din, N. (2018). Osteoid osteoma: Contemporary management. Orthopedic Reviews, 10(3), 108-119. Available at: https://ecommons.aku.edu/pakistan_fhs_mc_surg_orthop/92 Authors Shahryar Noordin, Salim Allana, Kiran Hilal, Riaz Hussain Lukhadwala, Anum Sadruddin Pidani, and Nasir Ud Din This article is available at eCommons@AKU: https://ecommons.aku.edu/pakistan_fhs_mc_surg_orthop/92 Orthopedic Reviews 2018; volume 10:7496 Osteoid osteoma: Contemporary management Epidemiology Correspondence: Shahryar Noordin, Orthopaedic Surgery, Aga Khan University, Osteoid osteoma accounts for around Karachi, Pakistan. Shahryar Noordin,1 Salim Allana,2 5% of all bone tumors and 11% of benign Tel.: 021.3486.4384. 4 Kiran Hilal,3 Naila Nadeem,3 bone tumors. Osteoid osteoma is the third E-mail: [email protected] Riaz Lakdawala,1 Anum Sadruddin,4 most common biopsy analyzed benign bone 5 tumor after osteochondroma and nonossify- Key words: Osteoid osteoma; tumor; benign; Nasir Uddin imaging; pathogenesis; management. 1 ing fibroma. Two to 3% of excised primary Orthopaedic Surgery, Aga Khan bone tumors are osteoid osteomas.5 Males University, Karachi, Pakistan; Contributions: SN, SA, study design, data col- are more commonly affected with an lection, manuscript writing; KH, NU, data col- 2 5 Department of Epidemiology, Rollins approximate male/female ratio of 2 to 1. -

View Presentation Notes
When is a musculoskeletal condition a tumor? Recognizing common bone and soft tissue tumors Christian M. Ogilvie, MD Assistant Professor of Orthopaedic Surgery University of Pennsylvania University of Pennsylvania Department of Orthopaedic Surgery Purpose • Recognize that tumors can present in the extremities of patients treated by athletic trainers • Know that tumors may present as a lump, pain or both • Become familiar with some bone and soft tissue tumors University of Pennsylvania Department of Orthopaedic Surgery Summary • Introduction – Pain – Lump • Bone tumors – Malignant – Benign • Soft tissue tumors – Malignant – Benign University of Pennsylvania Department of Orthopaedic Surgery Summary • Presentation • Imaging • History • Similar conditions –Injury University of Pennsylvania Department of Orthopaedic Surgery Introduction •Connective tissue tumors -Bone -Cartilage -Muscle -Fat -Synovium (lining of joints, tendons & bursae) -Nerve -Vessels •Malignant (cancerous): sarcoma •Benign University of Pennsylvania Department of Orthopaedic Surgery Introduction: Pain • Malignant bone tumors: usually • Benign bone tumors: some types • Malignant soft tissue tumors: not until large • Benign soft tissue tumors: some types University of Pennsylvania Department of Orthopaedic Surgery Introduction: Pain • Bone tumors – Not necessarily activity related – May be worse at night – Absence of trauma, mild trauma or remote trauma • Watch for referred patterns – Knee pain for hip problem – Arm and leg pains in spine lesions University of Pennsylvania -

Basal Cell Carcinoma Associated with Non-Neoplastic Cutaneous Conditions: a Comprehensive Review
Volume 27 Number 2| February 2021 Dermatology Online Journal || Review 27(2):1 Basal cell carcinoma associated with non-neoplastic cutaneous conditions: a comprehensive review Philip R Cohen MD1,2 Affiliations: 1San Diego Family Dermatology, National City, California, USA, 2Touro University California College of Osteopathic Medicine, Vallejo, California, USA Corresponding Author: Philip R Cohen, 10991 Twinleaf Court, San Diego, CA 92131-3643, Email: [email protected] pathogenesis of BCC is associated with the Abstract hedgehog signaling pathway and mutations in the Basal cell carcinoma (BCC) can be a component of a patched homologue 1 (PCTH-1) transmembrane collision tumor in which the skin cancer is present at tumor-suppressing protein [2-4]. Several potential the same cutaneous site as either a benign tumor or risk factors influence the development of BCC a malignant neoplasm. However, BCC can also concurrently occur at the same skin location as a non- including exposure to ultraviolet radiation, genetic neoplastic cutaneous condition. These include predisposition, genodermatoses, immunosuppression, autoimmune diseases (vitiligo), cutaneous disorders and trauma [5]. (Darier disease), dermal conditions (granuloma Basal cell carcinoma usually presents as an isolated faciale), dermal depositions (amyloid, calcinosis cutis, cutaneous focal mucinosis, osteoma cutis, and tumor on sun-exposed skin [6-9]. However, they can tattoo), dermatitis, miscellaneous conditions occur as collision tumors—referred to as BCC- (rhinophyma, sarcoidal reaction, and varicose veins), associated multiple skin neoplasms at one site scars, surgical sites, systemic diseases (sarcoidosis), (MUSK IN A NEST)—in which either a benign and/or systemic infections (leischmaniasis, leprosy and malignant neoplasm is associated with the BCC at lupus vulgaris), and ulcers. -

A Rare Case of Peripheral Primitive Neuroectodermal Tumor in Palate
Surgery and Rehabilitation Case Report ISSN: 2514-5959 A rare case of peripheral primitive neuroectodermal tumor in palate Rajul Ranka1*, Anuj Jain2, Amol Gadbail3 and Minal Chaudhary1 1Oral Pathology and Microbiology, Sharad Pawar Dental College and Hospital, Sawangi(M), Wardha, India 2Department of Trauma and Emergency Medicine, All India Institute of Medical Sciences, Bhopal, India 3Department of Dentistry, Indira Gandhi Government Medical College, Nagpur, India Abstract Primitive Neuroectodermal Tumor (PNET) is an aggressive round cell malignant tumor which belongs to Ewing’s Sarcoma family. Peripheral PNET is rare in head and neck region and even rare in palate with handful of cases reported till date. A case of Peripheral PNET of palate in a 32-year-old female pa tient along with its clinico-pathologic, radiologic and immunohistologic features as well as its management is reported here. We emphasize the need of histopathology and immunohistochemistry (IHC) for early diagnosis of such aggressive tumors to improve the prognosis of patient by providing timely management. Introduction Case report A neuroectodermal tumor is a tumor of the central or peripheral A thirty-two years old female patient of Indian origin reported nervous system. They are divided into two categories viz. Group to the Out-Patient Department of our institute with a complaint of (I) tumors, such as the pituitary adenomas and carcinoid tumors, painful ulcer on her palate since five months. The ulcer was gradually represent tumors that show predominantly epithelial differenti ation. progressive but has now suddenly increased in size since few days. It Group (II) tumors, which incorporate malignant melanoma, olfactory was associated with dull aching, intermittent and localized pain which neuroblastoma, Ewing’s sar coma (EWS) and primitive neuroectodermal aggravated on mastication and relieved with time. -

Osteoid Osteoma and Your Everyday Practice
n Review Article Instructions 1. Review the stated learning objectives at the beginning cme ARTICLE of the CME article and determine if these objectives match your individual learning needs. 2. Read the article carefully. Do not neglect the tables and other illustrative materials, as they have been selected to enhance your knowledge and understanding. 3. The following quiz questions have been designed to provide a useful link between the CME article in the issue Osteoid Osteoma and your everyday practice. Read each question, choose the correct answer, and record your answer on the CME Registration Form at the end of the quiz. Petros J. Boscainos, MD, FRCSEd; Gerard R. Cousins, MBChB, BSc(MedSci), MRCS; 4. Type or print your full name and address and your date of birth in the space provided on the CME Registration Form. Rajiv Kulshreshtha, MBBS, MRCS; T. Barry Oliver, MBChB, MRCP, FRCR; 5. Indicate the total time spent on the activity (reading article and completing quiz). Forms and quizzes cannot be Panayiotis J. Papagelopoulos, MD, DSc processed if this section is incomplete. All participants are required by the accreditation agency to attest to the time spent completing the activity. educational objectives 6. Complete the Evaluation portion of the CME Regi stration Form. Forms and quizzes cannot be processed if the Evaluation As a result of reading this article, physicians should be able to: portion is incomplete. The Evaluation portion of the CME Registration Form will be separated from the quiz upon receipt at ORTHOPEDICS. Your evaluation of this activity will in no way affect educational1.