Neuromedin U Is Overexpressed in Pancreatic Cancer and Increases Invasiveness Via the Hepatocyte Growth Factor C-Met Pathway
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effects of Endothelin-1 on the Membrane Potential and Slow Waves in Circular Smooth Muscle of Rat Gastric Antrum
J. Smooth Muscle Res. (2004) 40 (4 & 5): 199–210 199 Original Effects of endothelin-1 on the membrane potential and slow waves in circular smooth muscle of rat gastric antrum Kenro IMAEDA1, Takashi KATO1, Naotsuka OKAYAMA1, Seiji IMAI1, Makoto SASAKI1, Hiromi KATAOKA1, Takahiro NAKAZAWA1, Hirotaka OHARA1, Yoshihiko KITO2 and Makoto ITOH1 1Department of Internal Medicine and Bioregulation, 2Department of Physiology, Nagoya City University Graduate School of Medical Sciences, Nagoya 467-8601, Japan Abstract Electrophysiological effects of endothelin-1 (ET-1) on circular smooth muscle of rat gastric antrum were investigated by using intracellular membrane potential recording techniques. ET-1 (10 nM) caused an initial hyperpolarization of the membrane which was followed by a sustained depolarization. ET-1 also increased the frequency but not the amplitude of slow waves. In the presence of the endothelin type A (ETA) receptor antagonist, BQ123 (1 µM), ET-1 (10 nM) depolarized the membrane and increased the frequency of slow waves, but without the initial hyperpolarization. The selective endothelin type B (ETB) receptor agonist, sarafotoxin S6c (10 nM), also depolarized the membrane and increased the frequency of slow waves. In the presence of the ETB receptor antagonist, BQ788 (1 µM), ET-1 (10 nM) hyperpolarized the membrane. However, in the presence of BQ788, ET-1 caused neither the depolarization nor the increase in the frequency of the slow waves. The ET-1-induced hyperpolarization was completely abolished by apamin (0.1 µM). In the presence of apamin, ET-1 depolarized the membrane and increased the frequency of slow waves. The ET-1-induced depolarization was significantly attenuated by 4,4’-diisothiocyanatostilbene-2,2’-disulphonic acid (DIDS, 0.3 mM). -

ILC2 Activation by Protozoan Commensal Microbes
International Journal of Molecular Sciences Review ILC2 Activation by Protozoan Commensal Microbes Kyle Burrows 1 , Louis Ngai 1 , Flora Wong 1,2, David Won 1 and Arthur Mortha 1,* 1 University of Toronto, Department of Immunology, Toronto, ON M5S 1A8, Canada; [email protected] (K.B.) [email protected] (L.N.); fl[email protected] (F.W.); [email protected] (D.W.) 2 Ranomics, Inc. Toronto, ON M5G 1X5, Canada * Correspondence: [email protected] Received: 3 September 2019; Accepted: 27 September 2019; Published: 30 September 2019 Abstract: Group 2 innate lymphoid cells (ILC2s) are a member of the ILC family and are involved in protective and pathogenic type 2 responses. Recent research has highlighted their involvement in modulating tissue and immune homeostasis during health and disease and has uncovered critical signaling circuits. While interactions of ILC2s with the bacterial microbiome are rather sparse, other microbial members of our microbiome, including helminths and protozoans, reveal new and exciting mechanisms of tissue regulation by ILC2s. Here we summarize the current field on ILC2 activation by the tissue and immune environment and highlight particularly new intriguing pathways of ILC2 regulation by protozoan commensals in the intestinal tract. Keywords: ILC2; protozoa; Trichomonas; Tritrichomonas musculis; mucosal immunity; taste receptors; succinate; intestinal immunity; type 2 immunity; commensals 1. The ILC Lineage 1.1. The Family of Innate Lymphoid Cells Research over the last decade has redirected focus away from classical immune cell interactions within lymphoid tissues towards immunity within non-lymphoid tissues. Within these tissues, immune interactions involve local adaptation and rapid responses by tissue-resident immune cells. -

Neural Regulation of Interactions Between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells
Prime Archives in Immunology: 2nd Edition Book Chapter Neural Regulation of Interactions between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells Weiwei Chen1,2, Qiang Shu2 and Jie Fan1,3,4* 1Department of Surgery, University of Pittsburgh School of Medicine, USA 2The Children‟s Hospital, Zhejiang University School of Medicine, National Clinical Research Center for Child Health, China 3Research and Development, Veterans Affairs Pittsburgh Healthcare System, USA 4McGowan Institute for Regenerative Medicine, University of Pittsburgh, USA *Corresponding Author: Jie Fan, Department of Surgery, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA Published January 18, 2021 This Book Chapter is a republication of an article published by Jie Fan, et al. at Frontiers in Immunology in October 2020. (Chen W, Shu Q and Fan J (2020) Neural Regulation of Interactions Between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells. Front. Immunol. 11:576929. doi: 10.3389/fimmu.2020.576929) How to cite this book chapter: Weiwei Chen, Qiang Shu, Jie Fan. Neural Regulation of Interactions between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells. In: Ajmal Khan, Ahmed Al-Harrasi, editors. Prime Archives in Immunology: 2nd Edition. Hyderabad, India: Vide Leaf. 2021. 1 www.videleaf.com Prime Archives in Immunology: 2nd Edition © The Author(s) 2021. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License(http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Author Contributions: WC collected the data and drafted the manuscript. WC, QS, and JF conceived and designed the study. -

Guthrie Cdna Resource Center
cDNA Resource Center cDNA Resource Center Catalog cDNA Resource Center Missouri University of Science and Technology 400 W 11th Rolla, MO 65409 TEL: (573) 341-7610 FAX: (573) 341-7609 EMAIL: [email protected] www.cdna.org September, 2008 1 cDNA Resource Center Visit our web site for product updates 2 cDNA Resource Center The cDNA Resource Center The cDNA Resource Center is a service provided by the faculty of the Department of Biological Sciences of Missouri University of Science and Technology. The purpose of the cDNA Resource Center is to further scientific investigation by providing cDNA clones of human proteins involved in signal transduction processes. This is achieved by providing high quality clones for important signaling proteins in a timely manner. By high quality, we mean that the clones are • Sequence verified • Propagated in a versatile vector useful in bacterial and mammalian systems • Free of extraneous 3' and 5' untranslated regions • Expression verified (in most cases) by coupled in vitro transcription/translation assays • Available in wild-type, epitope-tagged and common mutant forms (e.g., constitutively- active or dominant negative) By timely, we mean that the clones are • Usually shipped within a day from when you place your order. Clones can be ordered from our web pages, by FAX or by phone. Within the United States, clones are shipped by overnight courier (FedEx); international orders are shipped International Priority (FedEx). The clones are supplied for research purposes only. Details on use of the material are included on the Material Transfer Agreement (page 3). Clones are distributed by agreement in Invitrogen's pcDNA3.1+ vector. -

Gent Forms of Metalloproteinases in Hydra
Cell Research (2002); 12(3-4):163-176 http://www.cell-research.com REVIEW Structure, expression, and developmental function of early diver- gent forms of metalloproteinases in Hydra 1 2 3 4 MICHAEL P SARRAS JR , LI YAN , ALEXEY LEONTOVICH , JIN SONG ZHANG 1 Department of Anatomy and Cell Biology University of Kansas Medical Center Kansas City, Kansas 66160- 7400, USA 2 Centocor, Malvern, PA 19355, USA 3 Department of Experimental Pathology, Mayo Clinic, Rochester, MN 55904, USA 4 Pharmaceutical Chemistry, University of Kansas, Lawrence, KS 66047, USA ABSTRACT Metalloproteinases have a critical role in a broad spectrum of cellular processes ranging from the breakdown of extracellular matrix to the processing of signal transduction-related proteins. These hydro- lytic functions underlie a variety of mechanisms related to developmental processes as well as disease states. Structural analysis of metalloproteinases from both invertebrate and vertebrate species indicates that these enzymes are highly conserved and arose early during metazoan evolution. In this regard, studies from various laboratories have reported that a number of classes of metalloproteinases are found in hydra, a member of Cnidaria, the second oldest of existing animal phyla. These studies demonstrate that the hydra genome contains at least three classes of metalloproteinases to include members of the 1) astacin class, 2) matrix metalloproteinase class, and 3) neprilysin class. Functional studies indicate that these metalloproteinases play diverse and important roles in hydra morphogenesis and cell differentiation as well as specialized functions in adult polyps. This article will review the structure, expression, and function of these metalloproteinases in hydra. Key words: Hydra, metalloproteinases, development, astacin, matrix metalloproteinases, endothelin. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors
Ecology and Evolutionary Biology 2021; 6(3): 53-77 http://www.sciencepublishinggroup.com/j/eeb doi: 10.11648/j.eeb.20210603.11 ISSN: 2575-3789 (Print); ISSN: 2575-3762 (Online) An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors Miguel Angel Fuertes*, Carlos Alonso Department of Microbiology, Centre for Molecular Biology “Severo Ochoa”, Spanish National Research Council and Autonomous University, Madrid, Spain Email address: *Corresponding author To cite this article: Miguel Angel Fuertes, Carlos Alonso. An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors. Ecology and Evolutionary Biology. Vol. 6, No. 3, 2021, pp. 53-77. doi: 10.11648/j.eeb.20210603.11 Received: April 24, 2021; Accepted: May 11, 2021; Published: July 13, 2021 Abstract: Capturing conserved patterns in genes and proteins is important for inferring phenotype prediction and evolutionary analysis. The study is focused on the conserved patterns of the G protein-coupled receptors, an important superfamily of receptors. Olfactory receptors represent more than 2% of our genome and constitute the largest family of G protein-coupled receptors, a key class of drug targets. As no crystallographic structures are available, mechanistic studies rely on the use of molecular dynamic modelling combined with site-directed mutagenesis data. In this paper, we hypothesized that human-mouse orthologs coding for G protein-coupled receptors maintain, at speciation events, shared compositional structures independent, to some extent, of their percent identity as reveals a method based in the categorization of nucleotide triplets by their gross composition. The data support the consistency of the hypothesis, showing in ortholog G protein-coupled receptors the presence of emergent shared compositional structures preserved at speciation events. -

G Protein‐Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: G protein-coupled receptors. British Journal of Pharmacology (2019) 176, S21–S141 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: G protein-coupled receptors Stephen PH Alexander1 , Arthur Christopoulos2 , Anthony P Davenport3 , Eamonn Kelly4, Alistair Mathie5 , John A Peters6 , Emma L Veale5 ,JaneFArmstrong7 , Elena Faccenda7 ,SimonDHarding7 ,AdamJPawson7 , Joanna L Sharman7 , Christopher Southan7 , Jamie A Davies7 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Monash Institute of Pharmaceutical Sciences and Department of Pharmacology, Monash University, Parkville, Victoria 3052, Australia 3Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK 4School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 5Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 6Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 7Centre for Discovery Brain Sciences, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. Although the Concise Guide represents approximately 400 pages, the material presented is substantially reduced compared to information and links presented on the website. -
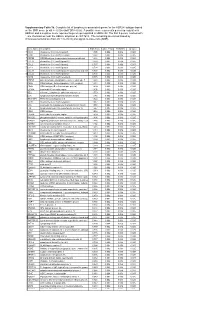
* Supplementary Table 3B. Complete List of Lymphocytic-Associated Genes
Supplementary Table 3b. Complete list of lymphocytic-associated genes for the HER2+I subtype based on the SNR score (p-val <= 0.005 and FDR<=0.05). A positive score represents genes up-regulated in HER2+I and a negative score represents genes up-regulated in HER2+NI. The first 9 genes, marked with *, are chemokines near the HER2+ amplicon at chr17q12. The remaining are sorted based by chromosomal location (from chr 1 to chr X) and signal-to-noise ratio (SNR). Gene Name Description SNR Score Feature P value FDR(BH) Q Value * CCL5 chemokine (C-C motif) ligand 5 1.395 0.002 0.036 0.025 * CCR7 chemokine (C-C motif) receptor 7 1.362 0.002 0.036 0.025 * CD79B CD79B antigen (immunoglobulin-associated beta) 1.248 0.002 0.036 0.025 * CCL13 chemokine (C-C motif) ligand 13 1.003 0.002 0.036 0.025 * CCL2 chemokine (C-C motif) ligand 2 0.737 0.004 0.056 0.037 * CCL8 chemokine (C-C motif) ligand 8 0.724 0.002 0.036 0.025 * CCL18 chemokine (C-C motif) ligand 18 (pulmonary and activ 0.703 0.002 0.036 0.025 * CCL23 chemokine (C-C motif) ligand 23 0.701 0.002 0.036 0.025 * CCR6 chemokine (C-C motif) receptor 6 0.715 0.002 0.036 0.025 PTPN7 protein tyrosine phosphatase, non-receptor type 7 1.861 0.002 0.036 0.025 CD3Z CD3Z antigen, zeta polypeptide (TiT3 complex) 1.811 0.002 0.036 0.025 CD48 CD48 antigen (B-cell membrane protein) 1.809 0.002 0.036 0.025 IL10RA interleukin 10 receptor, alpha 1.806 0.002 0.036 0.025 SELL selectin L (lymphocyte adhesion molecule 1) 1.773 0.002 0.036 0.025 LCK lymphocyte-specific protein tyrosine kinase 1.745 0.002 0.036 0.025 -

Neuromedins U and S Involvement in the Regulation of the Hypothalamo–Pituitary–Adrenal Axis
REVIEW ARTICLE published: 05 December 2012 doi: 10.3389/fendo.2012.00156 Neuromedins U and S involvement in the regulation of the hypothalamo–pituitary–adrenal axis Ludwik K. Malendowicz*, Agnieszka Ziolkowska and Marcin Rucinski Department of Histology and Embryology, Poznan University of Medical Sciences, Poznan, Poland Edited by: We reviewed neuromedin U (NMU) and neuromedin S (NMS) involvement in the regulation Hubert Vaudry, University of Rouen, of the hypothalamo–pituitary–adrenal (HPA) axis function. NMU and NMS are structurally France related and highly conserved neuropeptides. They exert biological effects via two GPCR Reviewed by: receptors designated as NMUR1 and NMUR2 which show differential expression. NMUR1 James A. Carr, Texas Tech University, USA is expressed predominantly at the periphery, while NMUR2 in the central nervous system. Gábor B. Makara, Hungarian Elements of the NMU/NMS and their receptors network are also expressed in the HPA Academy of Sciences, Hungary axis and progress in molecular biology techniques provided new information on their *Correspondence: actions within this system. Several lines of evidence suggest that within the HPA axis Ludwik K. Malendowicz, NMU and NMS act at both hypothalamic and adrenal levels. Moreover, new data suggest Department of Histology and Embryology, Poznan University of that NMU and NMS are involved in central and peripheral control of the stress response. Medical Sciences, 6 Swie¸cickiSt., 60-781 Poznan, Poland. Keywords: neuromedin U, neuromedin S, hypothalamus, pituitary, adrenal e-mail: [email protected] INTRODUCTION Identification of specific NMU receptors (NMUR1 and In search for new biologically active peptides, the group of NMUR2) and its anorexigenic action have enhanced interest in Minamino, Kangawa, and Matsuo in the 1980s isolated numerous physiological role of NMU and NMS (Howard et al., 2000; Ida small neuropeptides from porcine spinal cord. -

Proteins, Peptides, and Amino Acids
Proteins, Peptides, and Amino Acids Chandra Mohan, Ph.D. Calbiochem-Novabiochem Corp., San Diego, CA The Chemical Nature of Amino Acids Peptides and polypeptides are polymers of α-amino acids. There are 20 α-amino acids that make-up all proteins of biological interest. The α-amino acids in peptides and proteins α consist of a carboxylic acid (-COOH) and an amino (-NH2) functional group attached to the same tetrahedral carbon atom. This carbon is known as the -carbon. The type of R- group attached to this carbon distinguishes one amino acid from another. Several other amino acids, also found in the body, may not be associated with peptides or proteins. These non-protein-associated amino acids perform specialized functions. Some of the α-amino acids found in proteins are also involved in other functions in the body. For example, tyrosine is involved in the formation of thyroid hormones, and glutamate and aspartate act as neurotransmitters at fast junctions. R Amino acids exist in either D- or L- enantiomorphs or stereoisomers. The D- and L-refer to the absolute confirmation of optically active compounds. With the exception of glycine, all other amino acids are mirror images that can not be superimposed. Most of the amino acids found in nature are of the L-type. Hence, eukaryotic proteins are always composed of L-amino acids although D-amino acids are found in bacterial cell walls and in some peptide antibiotics. All biological reactions occur in an aqueous phase. Hence, it is important to know how the R-group of any given amino acid dictates the structure-function relationships of peptides and proteins in solution.