Mouse Relt Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Environmental Influences on Endothelial Gene Expression
ENDOTHELIAL CELL GENE EXPRESSION John Matthew Jeff Herbert Supervisors: Prof. Roy Bicknell and Dr. Victoria Heath PhD thesis University of Birmingham August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Tumour angiogenesis is a vital process in the pathology of tumour development and metastasis. Targeting markers of tumour endothelium provide a means of targeted destruction of a tumours oxygen and nutrient supply via destruction of tumour vasculature, which in turn ultimately leads to beneficial consequences to patients. Although current anti -angiogenic and vascular targeting strategies help patients, more potently in combination with chemo therapy, there is still a need for more tumour endothelial marker discoveries as current treatments have cardiovascular and other side effects. For the first time, the analyses of in-vivo biotinylation of an embryonic system is performed to obtain putative vascular targets. Also for the first time, deep sequencing is applied to freshly isolated tumour and normal endothelial cells from lung, colon and bladder tissues for the identification of pan-vascular-targets. Integration of the proteomic, deep sequencing, public cDNA libraries and microarrays, delivers 5,892 putative vascular targets to the science community. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Human RELT/TNFRSF19L Antibody Antigen Affinity-Purified Polyclonal Goat Igg Catalog Number: AF1385
Human RELT/TNFRSF19L Antibody Antigen Affinity-purified Polyclonal Goat IgG Catalog Number: AF1385 DESCRIPTION Species Reactivity Human Specificity Detects human RELT/TNFRSF19L in direct ELISAs and Western blots. In direct ELISAs, less than 1% crossreactivity with recombinant human (rh) 41BB, rhBAFF R, rhCD27, rhTAJ, rhCD30, rhDR3, rhDR6, rhTNF RI, rhTNF RII, rhEDAR, rhFas, rhGITR, rhHVEM, rhNGF R, rhOPG, and recombinant mouse OX40 is observed. Source Polyclonal Goat IgG Purification Antigen Affinitypurified Immunogen Mouse myeloma cell line NS0derived recombinant human RELT/TNFRSF19L Ser26Ala160 (Arg127Gly, Arg129Gly) Accession # Q969Z4 Formulation Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. See Certificate of Analysis for details. *Small pack size (SP) is supplied either lyophilized or as a 0.2 μm filtered solution in PBS. APPLICATIONS Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website. Recommended Sample Concentration Western Blot 1 µg/mL See Below DATA Western Blot Detection of Human RELT/TNFRSF19L by Western Blot. Western blot shows lysates of human bone marrow tissue and human lymph node tissue. PVDF membrane was probed with 1 µg/mL of Goat AntiHuman RELT/TNFRSF19L Antigen Affinitypurified Polyclonal Antibody (Catalog # AF1385) followed by HRPconjugated AntiGoat IgG Secondary Antibody (Catalog # HAF017). A specific band was detected for RELT/TNFRSF19L at approximately 50 kDa (as indicated). This experiment was conducted under reducing conditions and using Immunoblot Buffer Group 1. PREPARATION AND STORAGE Reconstitution Reconstitute at 0.2 mg/mL in sterile PBS. -

Produktinformation
Produktinformation Diagnostik & molekulare Diagnostik Laborgeräte & Service Zellkultur & Verbrauchsmaterial Forschungsprodukte & Biochemikalien Weitere Information auf den folgenden Seiten! See the following pages for more information! Lieferung & Zahlungsart Lieferung: frei Haus Bestellung auf Rechnung SZABO-SCANDIC Lieferung: € 10,- HandelsgmbH & Co KG Erstbestellung Vorauskassa Quellenstraße 110, A-1100 Wien T. +43(0)1 489 3961-0 Zuschläge F. +43(0)1 489 3961-7 [email protected] • Mindermengenzuschlag www.szabo-scandic.com • Trockeneiszuschlag • Gefahrgutzuschlag linkedin.com/company/szaboscandic • Expressversand facebook.com/szaboscandic SANTA CRUZ BIOTECHNOLOGY, INC. RELL1 siRNA (h): sc-156061 BACKGROUND PRODUCT RELL1, also known as RELT-like protein 1, is a 271 amino acid single-pass RELL1 siRNA (h) is a pool of 3 target-specific 19-25 nt siRNAs designed to type I membrane protein that belongs to the RELT family. RELL1 is widely knock down gene expression. Each vial contains 3.3 nmol of lyophilized expressed, with highest levels in the placenta, skeletal muscle, spleen and siRNA, sufficient for a 10 µM solution once resuspended using protocol testis. RELL1 shares 32% and 40% amino acid identity with RELT and RELL2, below. Suitable for 50-100 transfections. Also see RELL1 shRNA Plasmid (h): respectively. The RELL1 protein interacts with RELT, RELL2 and OXSR1, and sc-156061-SH and RELL1 shRNA (h) Lentiviral Particles: sc-156061-V as alter- is phosphorylated in vitro by OXSR1. The RELL1 gene is conserved in chim- nate gene silencing products. panzee, canine, bovine, mouse, rat and chicken, and maps to human chro- For independent verification of RELL1 (h) gene silencing results, we also pro- mosome 4p14. -

IMMUNOLOGY 2019™ Program
Scientific Program Book IMMUNOLOGY 2019™ The Annual Meeting of the American Association of Immunologists May 9–13, 2019 San Diego Convention Center San Diego, California TABLE OF CONTENTS Thursday Afternoon Sessions .......................................................................................... 1 Friday Morning Sessions .................................................................................................. 7 Friday Afternoon Sessions ............................................................................................... 13 Friday Posters ...................................................................................................................19 Saturday Morning Sessions ............................................................................................. 43 Saturday Afternoon Sessions ........................................................................................... 50 Saturday Posters .............................................................................................................. 56 Sunday Morning Sessions ............................................................................................... 82 Sunday Afternoon Sessions ............................................................................................. 88 Sunday Posters ................................................................................................................ 94 Monday Morning Sessions .............................................................................................. -

Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis
Getting “Under the Skin”: Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis by Kristen Monét Brown A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Epidemiological Science) in the University of Michigan 2017 Doctoral Committee: Professor Ana V. Diez-Roux, Co-Chair, Drexel University Professor Sharon R. Kardia, Co-Chair Professor Bhramar Mukherjee Assistant Professor Belinda Needham Assistant Professor Jennifer A. Smith © Kristen Monét Brown, 2017 [email protected] ORCID iD: 0000-0002-9955-0568 Dedication I dedicate this dissertation to my grandmother, Gertrude Delores Hampton. Nanny, no one wanted to see me become “Dr. Brown” more than you. I know that you are standing over the bannister of heaven smiling and beaming with pride. I love you more than my words could ever fully express. ii Acknowledgements First, I give honor to God, who is the head of my life. Truly, without Him, none of this would be possible. Countless times throughout this doctoral journey I have relied my favorite scripture, “And we know that all things work together for good, to them that love God, to them who are called according to His purpose (Romans 8:28).” Secondly, I acknowledge my parents, James and Marilyn Brown. From an early age, you two instilled in me the value of education and have been my biggest cheerleaders throughout my entire life. I thank you for your unconditional love, encouragement, sacrifices, and support. I would not be here today without you. I truly thank God that out of the all of the people in the world that He could have chosen to be my parents, that He chose the two of you. -

Systems Genetics Analysis of the LXS Recombinant Inbred Mouse Strains:Genetic and Molecular Insights Into Acute Ethanol Tolerance
PLOS ONE RESEARCH ARTICLE Systems genetics analysis of the LXS recombinant inbred mouse strains:Genetic and molecular insights into acute ethanol tolerance 1,2 3,4,5 3 3 Richard A. RadcliffeID *, Robin DowellID , Aaron T. Odell , Phillip A. RichmondID , 1 1 6 1 Beth Bennett , Colin LarsonID , Katerina KechrisID , Laura M. SabaID , 7 6 Pratyaydipta RudraID , Shi WenID a1111111111 1 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of Colorado Anschutz Medical Campus, Aurora, CO, United States of America, 2 Institute for Behavioral Genetics, University of Colorado a1111111111 Boulder, Boulder CO, United States of America, 3 BioFrontiers Institute, University of Colorado Boulder, a1111111111 Boulder, CO, United States of America, 4 Department of Molecular, Cellular, and Developmental Biology, a1111111111 University of Colorado Boulder, Boulder, CO, United States of America, 5 Department of Computer Science, a1111111111 University of Colorado Boulder, Boulder, CO, United States of America, 6 Department of Biostatistics and Informatics, Colorado School of Public Health, University of Colorado Anschutz Medical Campus, Aurora, CO, United States of America, 7 Department of Statistics, Oklahoma State University, Stillwater, OK, United States of America * [email protected] OPEN ACCESS Citation: Radcliffe RA, Dowell R, Odell AT, Richmond PA, Bennett B, Larson C, et al. (2020) Abstract Systems genetics analysis of the LXS recombinant inbred mouse strains:Genetic and molecular We have been using the Inbred Long- and Short-Sleep mouse strains (ILS, ISS) and a insights into acute ethanol tolerance. PLoS ONE recombinant inbred panel derived from them, the LXS, to investigate the genetic underpin- 15(10): e0240253. https://doi.org/10.1371/journal. -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -

UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title Cardiac Stretch-Induced Transcriptomic Changes are Axis-Dependent Permalink https://escholarship.org/uc/item/7m04f0b0 Author Buchholz, Kyle Stephen Publication Date 2016 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Cardiac Stretch-Induced Transcriptomic Changes are Axis-Dependent A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioengineering by Kyle Stephen Buchholz Committee in Charge: Professor Jeffrey Omens, Chair Professor Andrew McCulloch, Co-Chair Professor Ju Chen Professor Karen Christman Professor Robert Ross Professor Alexander Zambon 2016 Copyright Kyle Stephen Buchholz, 2016 All rights reserved Signature Page The Dissertation of Kyle Stephen Buchholz is approved and it is acceptable in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California, San Diego 2016 iii Dedication To my beautiful wife, Rhia. iv Table of Contents Signature Page ................................................................................................................... iii Dedication .......................................................................................................................... iv Table of Contents ................................................................................................................ v List of Figures ................................................................................................................... -
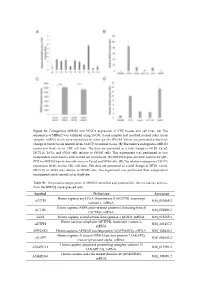
Figure S1. Endogenous MIR45
Figure S1. Endogenous MIR452 and VEGFA expression in CRC tissues and cell lines. (A) The expression of MIR452 was validated using 10 CRC tissue samples and matched normal colon tissue samples. miRNA levels were normalized to colon-specific RNU48. Values are presented as the fold- change in tumor tissue relative levels (ΔΔCT) to normal tissue. (B) The relative endogenous MIR452 expression levels in six CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, LoVo, and SW48 cells relative to SW480 cells. This experiment was performed as two independent experiments, each carried out in triplicate. (C) MIR452 expression level analysis by qRT- PCR for MIR452 transfection efficiency in Caco2 and SW48 cells. (D) The relative endogenous VEGFA expression levels in five CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, or SW48 cells relative to SW480 cells. This experiment was performed three independent experiments, each carried out in duplicate. Table S1. The putative target genes of MIR452 identified and predicted by the microarray analysis from the MIR452 overexpressed cells. Symbol Definition Accession Homo sapiens acyl-CoA thioesterase 8 (ACOT8), transcript ACOT8 NM_005469.2 variant 1, mRNA. Homo sapiens ARP6 actin-related protein 6 homolog (yeast) ACTR6 NM_022496.3 (ACTR6), mRNA. ADI1 Homo sapiens acireductone dioxygenase 1 (ADI1), mRNA. NM_018269.1 Homo sapiens aftiphilin (AFTPH), transcript variant 1, AFTPH NM_203437.2 mRNA. AHNAK2 Homo sapiens AHNAK nucleoprotein 2 (AHNAK2), mRNA. NM_138420.2 Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), AKAP7 NM_004842.2 transcript variant alpha, mRNA. Homo sapiens anaphase promoting complex subunit 13 ANAPC13 NM_015391.2 (ANAPC13), mRNA. -

Proteomic and Genomic Analysis of Acid Dentin Lysate with Focus on TGF
www.nature.com/scientificreports OPEN Proteomic and genomic analysis of acid dentin lysate with focus on TGF‑β signaling Jila Nasirzade1, Zahra Kargarpour1, Goran Mitulović2, Franz Josef Strauss1,3,4, Layla Panahipour1, Frank Schwarz5 & Reinhard Gruber1,6* Particulate autologous tooth roots are increasingly used for alveolar bone augmentation; however, the proteomic profle of acid dentin lysate and the respective cellular response have not been investigated. Here we show that TGF‑β1 is among the 226 proteins of acid dentin lysate (ADL) prepared from porcine teeth. RNA sequencing identifed 231 strongly regulated genes when gingival fbroblasts were exposed to ADL. Out of these genes, about one third required activation of the TGF‑β receptor type I kinase including interleukin 11 (IL11) and NADPH oxidase 4 (NOX4). Reverse transcription‑quantitative polymerase chain reaction and immunoassay confrmed the TGF‑β‑ dependent expression of IL11 and NOX4. The activation of canonical TGF‑β signaling by ADL was further confrmed by the phosphorylation of Smad3 and translocation of Smad2/3, using Western blot and immunofuorescence staining, respectively. Finally, we showed that TGF‑β activity released from dentin by acid lysis adsorbs to titanium and collagen membranes. These fndings suggest that dentin particles are a rich source of TGF‑β causing a major response of gingival fbroblasts. Autogenous tooth roots have brought a new approach to the feld of oral bone augmentation 1. Tooth roots were introduced due to the structural similarity between dentin and alveolar bone. Clinically, autogenous tooth roots have shown promising results in bone augmentation2 and prior to implant placement3 leading to similar outcomes compared to bone substitutes.