COPI Mediates Recycling of an Exocytic SNARE by Recognition of A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
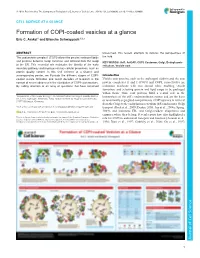
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

The Structure of the COPI Coat Determined Within the Cell
RESEARCH ADVANCE The structure of the COPI coat determined within the cell Yury S Bykov1,2, Miroslava Schaffer3, Svetlana O Dodonova1†, Sahradha Albert3, Ju¨ rgen M Plitzko3, Wolfgang Baumeister3*, Benjamin D Engel3*, John AG Briggs1,2* 1Structural and Computational Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany; 2Structural Studies Division, MRC Laboratory of Molecular Biology, Cambridge, United Kingdom; 3Department of Molecular Structural Biology, Max Planck Institute of Biochemistry, Martinsried, Germany Abstract COPI-coated vesicles mediate trafficking within the Golgi apparatus and from the Golgi to the endoplasmic reticulum. The structures of membrane protein coats, including COPI, have been extensively studied with in vitro reconstitution systems using purified components. Previously we have determined a complete structural model of the in vitro reconstituted COPI coat (Dodonova et al., 2017). Here, we applied cryo-focused ion beam milling, cryo-electron tomography and subtomogram averaging to determine the native structure of the COPI coat within vitrified Chlamydomonas reinhardtii cells. The native algal structure resembles the in vitro mammalian structure, but additionally reveals cargo bound beneath b’–COP. We find that all coat components *For correspondence: disassemble simultaneously and relatively rapidly after budding. Structural analysis in situ, [email protected] (WB); maintaining Golgi topology, shows that vesicles change their size, membrane thickness, and cargo [email protected] (BDE); content as they progress from cis to trans, but the structure of the coat machinery remains [email protected] constant. (JAGB) DOI: https://doi.org/10.7554/eLife.32493.001 Present address: †Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Go¨ ttingen, Germany Introduction Vesicular transport between cellular compartments is a fundamental mechanism of eukaryotic cells. -

The Amino-Terminal Domain of the Golgi Protein Giantin Interacts Directly with the Vesicle-Tethering Protein P115*
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 275, No. 4, Issue of January 28, pp. 2831–2836, 2000 © 2000 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. The Amino-terminal Domain of the Golgi Protein Giantin Interacts Directly with the Vesicle-tethering Protein p115* (Received for publication, November 10, 1999) Giovanni M. Lesa‡§¶, Joachim Seemann‡ʈ**, James Shorter‡ ‡‡, Joe¨l Vandekerckhove§§¶¶, and Graham Warren‡**ʈʈ From the ‡Cell Biology Laboratory and §Molecular Neuropathobiology Laboratory, Imperial Cancer Research Fund, 44 Lincoln’s Inn Fields, London, WC2A 3PX, United Kingdom and the §§Departments of Biochemistry and Medical Protein Research (VIB), Universiteit Gent, Ledeganckstraat 35, B-9000 Gent, Belgium Giantin is thought to form a complex with p115 and munc-18/sec1 group of proteins (22, 23). In vitro studies em- Golgi matrix protein 130, which is involved in the reas- ploying yeast vacuoles suggest that after SNARE pairing a sembly of Golgi cisternae and stacks at the end of mito- phosphatase-dependent (24) and a Ca2ϩ/calmodulin-dependent sis. The complex is involved in the tethering of coat step (25) are required for fusion. protomer I vesicles to Golgi membranes and the initial It has become increasingly clear that before SNARE pairing stacking of reforming cisternae. Here we show that the COP vesicles become tethered to target membranes. Tethering NH2-terminal 15% of Giantin suffices to bind p115 in is thought to be mediated by rod-like proteins with extensive vitro and in vivo and to block cell-free Golgi reassembly. coiled-coil (23, 26–28). -

Cryo-Tomography of Coat Complexes ISSN 2059-7983
research papers Visualizing membrane trafficking through the electron microscope: cryo-tomography of coat complexes ISSN 2059-7983 Evgenia A. Markova and Giulia Zanetti* Institute of Structural and Molecular Biology, Birkbeck College, Malet Street, London WC1E 7HX, England. Received 8 March 2019 *Correspondence e-mail: [email protected] Accepted 12 April 2019 Coat proteins mediate vesicular transport between intracellular compartments, Keywords: cryo-electron tomography; which is essential for the distribution of molecules within the eukaryotic cell. three-dimensional reconstruction; coat proteins; The global arrangement of coat proteins on the membrane is key to their vesicular transport; COPII; subtomogram function, and cryo-electron tomography and subtomogram averaging have been averaging. used to study membrane-bound coat proteins, providing crucial structural insight. This review outlines a workflow for the structural elucidation of coat proteins, incorporating recent developments in the collection and processing of cryo-electron tomography data. Recent work on coat protein I, coat protein II and retromer performed on in vitro reconstitutions or in situ is summarized. These studies have answered long-standing questions regarding the mechanisms of membrane binding, polymerization and assembly regulation of coat proteins. 1. Introduction Recent advances in cryo-electron microscopy (cryo-EM) have enabled numerous high-resolution studies which have addressed long-standing structural biology questions. The large body of cryo-EM work carried out for protein structure characterization has utilized single-particle electron micro- scopy (Cheng, 2018). Recently, developments in hardware, data collection and data processing have placed cryo-electron tomography (cryo-ET) and subtomogram averaging (STA) at the forefront of structural studies of repetitive assemblies, reaching resolutions comparable to those of single-particle EM (Schur et al., 2016; Wan et al., 2017; Hutchings et al., 2018; Dodonova et al., 2017; Himes & Zhang, 2018). -

June N&V Final
NEWS AND VIEWS studies is not fully functional, although this is disturbed, whereas binding of the calcium- our knowledge of the various steps in the not proven. An alternative explanation is that activated sytI domain would be sufficient to fusion process and their regulatory mecha- at synapses and in cells, full-length sytI (an promote SNARE zippering and fusion (Fig. 2). nisms. This might ultimately result in the integral membrane protein) is needed not only In conclusion, the results from Tucker et identification of other factors that regulate the for exocytic fusion, but also to initiate vesicle al.1 are an important milestone in the recon- system, in particular those allowing mem- docking to the plasma membrane through stitution of presynaptic events that result in brane fusion to operate in the sub-millisecond binding to SNAP25. In this case, inhibition by neurotransmitter release. On the basis of these timescale as it does in synaptic physiology. the cytosolic domain could be explained by an results, the need for proteins and lipids other 1. Tucker, W. C., Weber, T. & Chapman, E. R. Science unproductive interaction with the t-SNARE than sytI in the calcium regulation of synaptic 304, 435–438 (2004). 5,6 that prevents binding of full-length sytI (refs 9, vesicle fusion can finally be excluded. 2. Weber, T. et al. Cell 92, 759–72 (1998). 10). The reconstituted bilayers, however, could Exocytic fusion, however, is a complex process 3. Geppert, M. et al. Cell 79, 717–727 (1994). 4. Ubach, J. et al. EMBO J. 17, 3921–3930 (1998). -

Vesicle Coats: Structure, Function, And
Review Vesicle coats: structure, function, and general principles of assembly 1 2 2 1,3 Marco Faini , Rainer Beck , Felix T. Wieland , and John A.G. Briggs 1 Structural and Computational Biology Unit, European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany 2 Heidelberg University Biochemistry Center, Heidelberg University, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany 3 Cell Biology and Biophysics Unit, European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany The transport of proteins and lipids between distinct The three best-characterized types of vesicular carrier cellular compartments is conducted by coated vesicles. involved in intracellular trafficking are distinguished by These vesicles are formed by the self-assembly of coat their different coat proteins and their different trafficking proteins on a membrane, leading to collection of the routes. Clathrin-coated vesicles (CCVs) act in the late vesicle cargo and membrane bending to form a bud. secretory pathway and in the endocytic pathway, Scission at the bud neck releases the vesicle. X-ray COPII-coated vesicles export proteins from the endoplas- crystallography and electron microscopy (EM) have re- mic reticulum (ER), and COPI-coated vesicles shuttle cently generated models of isolated coat components within the Golgi organelle and from the Golgi back to and assembled coats. Here, we review these data to the ER. Despite having different compartment specifici- present a structural overview of the three main coats: ties and different structural components, the mechanisms clathrin, COPII, and COPI. The three coats have similar of their formation follow similar rules. The time and place function, common ancestry, and structural similarities, at which vesicle formation occurs are most often regulated but exhibit fundamental differences in structure and by small GTP-binding proteins. -

1 Novel Functions of the Flippase Drs2 and the Coat Protein COPI In
Novel Functions of the Flippase Drs2 and the Coat Protein COPI in Protein Trafficking By Hannah Marcille Hankins Dissertation Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biological Sciences May, 2016 Nashville, Tennessee Approved: Todd R. Graham, Ph.D Katherine L. Friedman, Ph.D. Kendal S. Broadie, Ph.D. Anne K. Kenworthy, Ph.D. Charles R. Sanders, Ph.D. 1 ACKNOWLEDGEMENTS It takes a village to raise a graduate student, and I have many villagers to thank for getting me to this point. I would never have considered doing research if it were not for my professors at Henderson State University, particularly Martin Campbell and Troy Bray, who were very supportive and encouraging throughout my undergraduate studies. I was also fortunate to be able to do full time research during the summers of my sophomore and junior years in the lab of Kevin Raney at the University of Arkansas for Medical Sciences. If it were not for the support system and these wonderful research experiences I had as an undergraduate, I never would have pursued graduate school at Vanderbilt and would not be the scientist I am today. I would like to thank my graduate school mentor, Todd Graham, for giving me the opportunity to join his lab and for introducing me to the wonderful world of yeast. Todd is an amazing and enthusiastic scientist; his guidance and support throughout my graduate career has been invaluable and I am truly grateful to have such a wonderful mentor. -

Three Ways to Make a Vesicle
REVIEWS THREE WAYS TO MAKE A VESICLE Tomas Kirchhausen Cargo molecules have to be included in carrier vesicles of different forms and sizes to be transported between organelles. During this process, a limited set of proteins, including the coat proteins COPI, COPII and clathrin, carries out a programmed set of sequential interactions that lead to the budding of vesicles. A general model to explain the formation of coated vesicles is starting to emerge but the picture is more complex than we had imagined. ENDOCYTIC PATHWAY Transport of proteins and lipids along the ENDOCYTIC or energy dependent and includes sorting of cargo to the Macromolecules are taken up SECRETORY PATHWAYS is a hallmark of eukaryotic cells. The forming coat. Coat propagation, the second step in the by invagination of the plasma membrane fluxes along these pathways are very large process, couples further addition of coat components membrane. They first arrive in and rapid. A FIBROBLAST kept in resting conditions in a and additional recruitment of cargo with invagination of early endosomes, then late endosomes, and finally tissue culture plate internalizes an amount of mem- the underlying membrane. When formation of the coat lysosomes, where they are brane equivalent to the whole surface area of the cell in ends, the vesicle buds by scission of the neck connecting degraded by hydrolases. one hour. Inside the cell, it often takes only seconds for a the deeply invaginated membrane to the donor surface. carrier vesicle to move from the donor membrane to This is a relatively simple step for COPI- and COPII- SECRETORY PATHWAY the acceptor organelle. -

Structure of Coatomer Cage Proteins and the Relationship Among COPI
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector StructureofCoatomerCageProteins and the Relationship among COPI, COPII, and Clathrin Vesicle Coats Changwook Lee1,2 and Jonathan Goldberg1,2,* 1Howard Hughes Medical Institute 2Structural Biology Program Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10065, USA *Correspondence: [email protected] DOI 10.1016/j.cell.2010.05.030 SUMMARY of GDP for GTP on ARF catalyzed by a Golgi-localized guanine- nucleotide exchange factor (GEF) of the Sec7 family (Peyroche COPI-coated vesicles form at the Golgi apparatus et al., 1996; Chardin et al., 1996). ARF-GTP binds to the from two cytosolic components, ARF G protein and membrane by embedding an N-terminal a helix in the bilayer coatomer, a heptameric complex that can poly- and in turn recruits coatomer through a direct, GTP-dependent merize into a cage to deform the membrane into interaction (Antonny et al., 1997; Zhao et al., 1997). Coatomer a bud. Although coatomer shares a common evolu- complexes, attached to the membrane surface through ARF- tionary origin with COPII and clathrin vesicle coat GTP, can bind cargo molecules and then self-assemble to form spherical cages that yield COPI-coated vesicles (Bremser proteins, the architectural relationship among the 0 et al., 1999; Nickel et al., 1998; Orcl et al., 1993; Spang et al., three cages is unclear. Strikingly, the ab -COP core 1998). of coatomer crystallizes as a triskelion in which three The architecture and functional organization of clathrin and 0 copies of a b -COP b-propeller domain converge COPII vesicle coats have been studied extensively by elec- through their axial ends. -

Retrograde Trafficking and Quality Control of Yeast Synaptobrevin, Snc1, Are Conferred by Its Transmembrane Domain
M BoC | ARTICLE Retrograde trafficking and quality control of yeast synaptobrevin, Snc1, are conferred by its transmembrane domain Mengxiao Ma and Christopher G. Burd* Department of Cell Biology, Yale School of Medicine, New Haven, CT 06520 ABSTRACT Synaptobrevin/vesicle-associated membrane protein 2 (VAMP2) is an essential Monitoring Editor soluble N-ethyl maleimide–sensitive factor attachment protein receptor (SNARE) protein that Patrick J. Brennwald has been extensively studied in its role in synaptic vesicle fusion. However, sorting and University of North Carolina trafficking of VAMP2 within the endosomal system is not well understood. Here, we use the Received: Feb 25, 2019 yeast VAMP2 homologue Snc1 to investigate the pathways and signals required for endocytic Revised: Apr 11, 2019 trafficking. We identify two genetically distinct retrieval pathways from the endosomal Accepted: May 3, 2019 system: a plasma membrane recycling pathway that requires the Rcy1 F-box protein and a retrograde pathway originating from the multivesicular/prevacuole endosome dependent on the Snx4-Atg20 sorting nexin complex. Lysine residues within the transmembrane domain of Snc1 are necessary for presentation of a Snx4-Atg20–dependent sorting signal located within its juxtamembrane region. Mutations of the transmembrane lysine residues ablate retrograde sorting and subject Snc1 to quality control via sorting into the degradative multivesicular endosome pathway. Degradative sorting requires lysine residues in the juxtamembrane region of Snc1 and is mediated by the Rsp5 ubiquitin ligase and its transmembrane adapters, Ear1 and Ssh4, which localize to endosome and vacuole membranes. This study shows that Snc1 is trafficked between the endosomal system and the Golgi apparatus via multiple pathways and provides evidence for protein quality control surveillance of a SNARE protein in the endo-vacuolar system. -

Coated Vesicles in Plant Cells Matthew J
Seminars in Cell & Developmental Biology 18 (2007) 471–478 Review Coated vesicles in plant cells Matthew J. Paul, Lorenzo Frigerio ∗ Department of Biological Sciences, University of Warwick, Coventry CV4 7AL, United Kingdom Available online 10 July 2007 Abstract Coated vesicles represent vital transport intermediates in all eukaryotic cells. While the basic mechanisms of membrane exchange are conserved through the kingdoms, the unique topology of the plant endomembrane system is mirrored by several differences in the genesis, function and regulation of coated vesicles. Efforts to unravel the complex network of proteins underlying the behaviour of these vesicles have recently benefited from the application in planta of several molecular tools used in mammalian systems, as well as from advances in imaging technology and the ongoing analysis of the Arabidopsis genome. In this review, we provide an overview of the roles of coated vesicles in plant cells and highlight salient new developments in the field. © 2007 Elsevier Ltd. All rights reserved. Keywords: Plant secretory pathway; Protein trafficking; Endocytosis; Endoplasmic reticulum; Golgi; Vacuole; Coated vesicles; Clathrin; COPI; COPII; Retromer Contents 1. Vesicular trafficking in plant cells ......................................................................................... 471 2. Small GTP-binding proteins and associated factors ......................................................................... 473 3. Plant vesicle coat families ............................................................................................... -

Modification of Intracellular Membrane Structures for Virus Replication
REVIEWS Modification of intracellular membrane structures for virus replication Sven Miller* and Jacomine Krijnse-Locker‡ Abstract | Viruses are intracellular parasites that use the host cell they infect to produce new infectious progeny. Distinct steps of the virus life cycle occur in association with the cytoskeleton or cytoplasmic membranes, which are often modified during infection. Plus- stranded RNA viruses induce membrane proliferations that support the replication of their genomes. Similarly, cytoplasmic replication of some DNA viruses occurs in association with modified cellular membranes. We describe how viruses modify intracellular membranes, highlight similarities between the structures that are induced by viruses of different families and discuss how these structures could be formed. Coatomer Viruses are small, obligatory-intracellular parasites structures involves interplay between the virus and the A coat complex that functions that contain either DNA or RNA as their genetic mate- host cell, the role of both viral and cellular proteins is in anterograde and retrograde rial. They depend entirely on host cells to replicate addressed. transport between the their genomes and produce infectious progeny. Viral endoplasmic reticulum and the penetration into the host cell is followed by genome Viruses and membranes Golgi apparatus. uncoating, genome expression and replication, assem- The cellular players. Cells are equipped with two major Clathrin bly of new virions and their egress. These steps can trafficking pathways to secrete and internalize material: First vesicle coat protein to be occur in close association with cellular structures, in the secretory and endocytic pathways (FIG. 1). identified; involved in particular cellular membranes and the cytoskeleton. Proteins that are destined for the extracellular environ- membrane trafficking to, and through, the endocytic Viruses are known to manipulate cells to facilitate their ment enter the secretory pathway upon co-translational pathway.