Brainomics: Harnessing the Cubicweb Semantic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
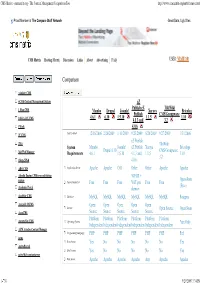
CMS Matrix - Cmsmatrix.Org - the Content Management Comparison Tool
CMS Matrix - cmsmatrix.org - The Content Management Comparison Tool http://www.cmsmatrix.org/matrix/cms-matrix Proud Member of The Compare Stuff Network Great Data, Ugly Sites CMS Matrix Hosting Matrix Discussion Links About Advertising FAQ USER: VISITOR Compare Search Return to Matrix Comparison <sitekit> CMS +CMS Content Management System eZ Publish eZ TikiWiki 1 Man CMS Mambo Drupal Joomla! Xaraya Bricolage Publish CMS/Groupware 4.6.1 6.10 1.5.10 1.1.5 1.10 1024 AJAX CMS 4.1.3 and 3.2 1Work 4.0.6 2F CMS Last Updated 12/16/2006 2/26/2009 1/11/2009 9/23/2009 8/20/2009 9/27/2009 1/31/2006 eZ Publish 2flex TikiWiki System Mambo Joomla! eZ Publish Xaraya Bricolage Drupal 6.10 CMS/Groupware 360 Web Manager Requirements 4.6.1 1.5.10 4.1.3 and 1.1.5 1.10 3.2 4Steps2Web 4.0.6 ABO.CMS Application Server Apache Apache CGI Other Other Apache Apache Absolut Engine CMS/news publishing 30EUR + system Open-Source Approximate Cost Free Free Free VAT per Free Free (Free) Academic Portal domain AccelSite CMS Database MySQL MySQL MySQL MySQL MySQL MySQL Postgres Accessify WCMS Open Open Open Open Open License Open Source Open Source AccuCMS Source Source Source Source Source Platform Platform Platform Platform Platform Platform Accura Site CMS Operating System *nix Only Independent Independent Independent Independent Independent Independent ACM Ariadne Content Manager Programming Language PHP PHP PHP PHP PHP PHP Perl acms Root Access Yes No No No No No Yes ActivePortail Shell Access Yes No No No No No Yes activeWeb contentserver Web Server Apache Apache -

A Framework for Ontology-Based Library Data Generation, Access and Exploitation
Universidad Politécnica de Madrid Departamento de Inteligencia Artificial DOCTORADO EN INTELIGENCIA ARTIFICIAL A framework for ontology-based library data generation, access and exploitation Doctoral Dissertation of: Daniel Vila-Suero Advisors: Prof. Asunción Gómez-Pérez Dr. Jorge Gracia 2 i To Adelina, Gustavo, Pablo and Amélie Madrid, July 2016 ii Abstract Historically, libraries have been responsible for storing, preserving, cata- loguing and making available to the public large collections of information re- sources. In order to classify and organize these collections, the library commu- nity has developed several standards for the production, storage and communica- tion of data describing different aspects of library knowledge assets. However, as we will argue in this thesis, most of the current practices and standards available are limited in their ability to integrate library data within the largest information network ever created: the World Wide Web (WWW). This thesis aims at providing theoretical foundations and technical solutions to tackle some of the challenges in bridging the gap between these two areas: library science and technologies, and the Web of Data. The investigation of these aspects has been tackled with a combination of theoretical, technological and empirical approaches. Moreover, the research presented in this thesis has been largely applied and deployed to sustain a large online data service of the National Library of Spain: datos.bne.es. Specifically, this thesis proposes and eval- uates several constructs, languages, models and methods with the objective of transforming and publishing library catalogue data using semantic technologies and ontologies. In this thesis, we introduce marimba-framework, an ontology- based library data framework, that encompasses these constructs, languages, mod- els and methods. -

Flemish Public Libraries – Digital Library Systems Architecture Study 10-Dec-13
Flemish Public Libraries – Digital Library Systems Architecture Study 10-Dec-13 Flemish Public Libraries - Digital Library Systems Architecture Study Last update: 27 NOV 2013 Researchers: Jean-François Declercq Rosemie Callewaert François Vermaut Ordered by: Bibnet vzw Vereniging van de Vlaamse Provincies Vlaamse Gemeenschapscommissie van het Brussels Hoofdstedelijk Gewest 2013-11-27-DigitalLibrarySystemArchitecture p. 1 / 163 Flemish Public Libraries – Digital Library Systems Architecture Study 10-Dec-13 Table of contents 1 Introduction ............................................................................................................................... 6 1.1 Definition of a Digital Library ............................................................................................. 6 1.2 Study Objectives ................................................................................................................ 7 1.3 Study Approach ................................................................................................................. 7 1.4 Study timeline .................................................................................................................... 8 1.5 Study Steering Committee ................................................................................................. 9 2 ICT Systems Architecture Concepts ......................................................................................... 10 2.1 Enterprise Architecture Model ....................................................................................... -

Download Slide (PDF Document)
When Django is too bloated Specialized Web-Applications with Werkzeug EuroPython 2017 – Rimini, Italy Niklas Meinzer @NiklasMM Gotthard Base Tunnel Photographer: Patrick Neumann Python is amazing for web developers! ● Bottle ● BlueBream ● CherryPy ● CubicWeb ● Grok ● Nagare ● Pyjs ● Pylons ● TACTIC ● Tornado ● TurboGears ● web2py ● Webware ● Zope 2 Why would I want to use less? ● Learn how stuff works Why would I want to use less? ● Avoid over-engineering – Wastes time and resources – Makes updates harder – It’s a security risk. Why would I want to use less? ● You want to do something very specific ● Plan, manage and document chemotherapy treatments ● Built with modern web technology ● Used by hospitals in three European countries Patient Data Lab Data HL7 REST Pharmacy System Database Printers Werkzeug = German for “tool” ● Developed by pocoo team @ pocoo.org – Flask, Sphinx, Jinja2 ● A “WSGI utility” ● Very lightweight ● No ORM, No templating engine, etc ● The basis of Flask and others Werkzeug Features Overview ● WSGI – WSGI 1.0 compatible, WSGI Helpers ● Wrapping of requests and responses ● HTTP Utilities – Header processing, form data parsing, cookies ● Unicode support ● URL routing system ● Testing tools – Testclient, Environment builder ● Interactive Debugger in the Browser A simple Application A simple Application URL Routing Middlewares ● Separate parts of the Application as wsgi apps ● Combine as needed Request Static files DB Part of Application conn with DB access User Dispatcher auth Part of Application without DB access Response HTTP Utilities ● Work with HTTP dates ● Read and dump cookies ● Parse form data Using the test client Using the test client - pytest fixtures Using the test client - pytest fixtures Interactive debugger in the Browser Endless possibilities ● Connect to a database with SQLalchemy ● Use Jinja2 to render documents ● Use Celery to schedule asynchronous tasks ● Talk to 3rd party APIs with requests ● Make syscalls ● Remote control a robot to perform tasks at home Thank you! @NiklasMM NiklasMM Photographer: Patrick Neumann. -

Application from the Bibliothèque Nationale De France (Bnf) for Gallica (Gallica.Bnf.Fr) and Data (Data.Bnf.Fr) TABLE of CONTENTS I
Stanford Prize for Innovation in Research Libraries (SPIRL) Application from the Bibliothèque nationale de France (BnF) for Gallica (gallica.bnf.fr) and Data (data.bnf.fr) TABLE OF CONTENTS I. Nominators’ statement ........................................................................................................... 1 II. Narrative description ............................................................................................................ 2 1. THE GALLICA DIGITAL LIBRARY: AN ONGOING INNOVATION .................................... 2 1.1 A BRIEF HISTORY OF GALLICA ..................................................................................... 2 Gallica today and tomorrow ................................................................................................... 6 1.2 GALLICA TRULY CARES ABOUT ITS END USERS ....................................................... 6 Smart search .......................................................................................................................... 6 Smart browsing ...................................................................................................................... 7 Gallica made mobile .............................................................................................................. 7 1.3 A LIBRARY DESIGNED FOR USE AND RE-USE ............................................................ 8 Terms of use .......................................................................................................................... 8 Off line -

Brainomics: Harnessing the Cubicweb Semantic
Brainomics: Harnessing the CubicWeb semantic framework to manage large neuromaging genetics shared resources David Goyard, Antoine Grigis, Dimitri Papadopoulos-Orfanos, Michel Vittot, Vincent Frouin, Adrien Di Mascio To cite this version: David Goyard, Antoine Grigis, Dimitri Papadopoulos-Orfanos, Michel Vittot, Vincent Frouin, et al.. Brainomics: Harnessing the CubicWeb semantic framework to manage large neuromaging genetics shared resources. Journées RITS 2015, Mar 2015, Dourdan, France. p34-35 Section imagerie génétique. inserm-01145600 HAL Id: inserm-01145600 https://www.hal.inserm.fr/inserm-01145600 Submitted on 24 Apr 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Actes des Journées Recherche en Imagerie et Technologies pour la Santé - RITS 2015 34 Brainomics: Harnessing the CubicWeb semantic framework to manage large neuromaging genetics shared resources 1 1 1 2 1 2 D. Goyard , A. Grigis , D. Papadopoulos Orfanos , V. Michel , V. Frouin ∗, A. Di Mascio 1 CEA DSV NeuroSpin, UNATI, 91 Gif sur Yvette, France. 2 Logilab, Paris, France. ∗ [email protected]. Abstract - In neurosciences or psychiatry, large mul- one workpackage devoted to this issue. The package was ticentric population studies are being acquired and the twofolds: (i) to develop specific Python modules using the corresponding data are made available to the acquisi- CubicWeb framework (Logilab, Paris) and (ii) to demon- tion partners or the scientific community. -

Efficient Querying of Distributed Sources in a Mobile Environment Through Source Indexing and Caching
Faculty of Science, Department of Computer science Efficient querying of distributed sources in a mobile environment through source indexing and caching. Graduation thesis submitted in partial fulfillment of the requirements for the degree of Master in Applied Informatics. Elien Paret Promoter: Prof. Dr. Olga De Troyer Advisors: Dr. Sven Casteleyn & William Van Woensel Academic year 2009 -2010 Acknowledgements In this small chapter I would like to thank everyone that helped me achieve this thesis. First of all I would like to thank my promoter, Prof. Olga De Troyer, for providing me the opportunity to realize this thesis and for her personal assistance during this entire academic year. Secondly, I would like to express my deepest gratitude to my two excellent advisors, Dr. Sven Casteleyn and PhD William Van Woensel. I am very thankful for their personal guidance, moral support and all the knowledge they shared with me. I also want to thank my dear boyfriend Pieter Callewaert for his moral support, kindness, patience, encouragement and for being there for me at all circumstances and times. Finally I would like to thank my family for their love and support during my entire life and for supporting me getting my masters degree and achieving this thesis. Thank you all. -i- Abstract Mobile devices have become a part of the everyday life; they are used anywhere and at anytime, for communication, looking up information, consulting an agenda, making notes, playing games etc. At the same time, the hardware of these devices has evolved significantly: e.g., faster processors, larger memory and improved connectivity … The hardware evolution along with the recent advancements of identification techniques, has lead to new opportunities for developers of mobile applications: mobile applications can be aware of their environment and the objects in it. -

Isa2 Action 2017.01 Standard-Based Archival
Ref. Ares(2018)3256671 - 20/06/2018 ISA2 ACTION 2017.01 STANDARD-BASED ARCHIVAL DATA MANAGEMENT, EXCHANGE AND PUBLICATION STUDY FINAL REPORT Study on Standard-Based Archival Data Management, Exchange and Publication Final Report DOCUMENT METADATA Property Value Release date 15/06/2018 Status: Final version Version: V1.00 Susana Segura, Luis Gallego, Emmanuel Jamin, Miguel Angel Gomez, Seth Authors: van Hooland, Cédric Genin Lieven Baert, Julie Urbain, Annemie Vanlaer, Belá Harsanyi, Razvan Reviewed by: Ionescu, Reflection Committee Approved by: DOCUMENT HISTORY Version Description Action 0.10 First draft 0.90 Version for Review 0.98 Second version for Review 0.99 Third version for Acceptance 1.00 Final version 2 Study on Standard-Based Archival Data Management, Exchange and Publication Final Report TABLE OF CONTENTS Table of Contents ........................................................................................................................ 3 List of Figures ............................................................................................................................. 8 List of Tables ............................................................................................................................. 10 1 Executive Summary ........................................................................................................... 14 2 Introduction ........................................................................................................................ 16 2.1 Context .......................................................................................................................... -

Publishing Bibliographic Records on the Web of Data: Opportunities for the Bnf (French National Library)
Publishing Bibliographic Records on the Web of Data: Opportunities for the BnF (French National Library) Agnès Simon1, Romain Wenz1, Vincent Michel2, Adrien Di Mascio2 1 Bibliothèque nationale de France, Paris, France [email protected], [email protected] 2 Logilab, Paris, France [email protected], [email protected] Abstract. Linked open data tools have been implemented through data.bnf.fr, a project which aims at making the BnF data more useful on the Web. data.bnf.fr gathers data automatically from different databases on pages about authors, works and themes. Online since July 2011, it is still under development and has feed- backs from several users, already. First the article will present the issues linked to our data and stress the importance of useful links and of persistency for archival purposes. We will discuss our so- lution and methodology, showing their strengths and weaknesses, to create new services for the library. An insight on the ontology and vocabularies will be given, with a “business” view of the interaction between rich RDF ontologies and light HTML embedded data such as schema.org. The broader question of Libraries on the Semantic Web will be addressed so as to help specify similar projects. Keywords: Libraries, Open data, Culture, Project management, Entity linking, Relational database, CubicWeb, Encoded Archival Description, Marc formats, Open Archive Initiative, Text Encoding Initiative. 1 Introduction The BnF (French national library) sees Semantic Web technologies as an opportunity to weave its data into the Web and to bring structure and reliability to existing information. The BnF is one of the most important heritage institutions in France, with a history going back to the 14th century and millions of documents, including a large variety of hand-written, printed and digital material, through millions of bibliographic records. -

Produire Des Contenus Documentaires En Ligne Quelles Stratégies Pour Les Bibliothèques ?
Produire des contenus documentaires en ligne Quelles stratégies pour les bibliothèques ? Christelle Di Pietro (dir.) DOI : 10.4000/books.pressesenssib.2814 Éditeur : Presses de l’enssib Lieu d'édition : Villeurbanne Année d'édition : 2014 Date de mise en ligne : 10 décembre 2018 Collection : La Boîte à outils ISBN électronique : 9782375460603 http://books.openedition.org Édition imprimée Date de publication : 1 janvier 2014 ISBN : 9791091281379 Nombre de pages : 188 Référence électronique DI PIETRO, Christelle (dir.). Produire des contenus documentaires en ligne : Quelles stratégies pour les bibliothèques ? Nouvelle édition [en ligne]. Villeurbanne : Presses de l’enssib, 2014 (généré le 03 mai 2019). Disponible sur Internet : <http://books.openedition.org/pressesenssib/2814>. ISBN : 9782375460603. DOI : 10.4000/books.pressesenssib.2814. © Presses de l’enssib, 2014 Conditions d’utilisation : http://www.openedition.org/6540 #30 PRODUIRE DES CONTENUS DOCUMENTAIRES EN LIGNE : QUELLES STRATÉGIES POUR LES BIBLIOTHÈQUES ? sous la direction de Christelle Di Pietro 30 BAO#30 PRODUIRE DES CONTENUS DOCUMENTAIRES EN LIGNE : QUELLES STRATÉGIES POUR LES BIBLIOTHÈQUES ? +++++++++++++++++++++++++++++++++++++++++++++++++++++++++ Plébiscités par les publics, les services d’information se multiplient et se diversiient. La gestion des collections en bibliothèque implique désormais leur exploitation et la réappropriation de ces contenus pour élaborer toute une gamme de produits documentaires. Nouvelles compétences du professionnel, renouvellement des formes de collaboration, aussi bien sur les aspects techniques, juridiques que métho- dologiques et rédactionnels, ce volume investit de façon inédite le champ de l’Internet de contenus pour tous les types de publics. Le plan de l’ouvrage s’articule autour de quatre parties : exploiter les collec- tions et repenser les accès en ligne, les produits documentaires de synthèse : curation et production, produire en co-construction et en réseau, et enin, les outils et le droit. -

Python As a Tool for Web Server Application Development Sheetal Taneja1, Pratibha R
JIMS 8i-International Journal of Information, Communication and Computing Technology(IJICCT) Python as a Tool for Web Server Application Development Sheetal Taneja1, Pratibha R. Gupta2 specific request, there is a program running at the server end ABSTRACT that performs this task. Such a program is termed Web application [2]. Thus, web applications are the programs With evolution of web, several competitive languages such running at application server and are invoked using browser as Java, PHP, Python, Ruby are catching the attention of the through the Internet. developers. Recently Python has emerged as a popular and the preferred web programming language, because of its Web applications offer several benefits over traditional simplicity to code and ease of learning. Being a flexible applications that are required to be installed at each host language, it offers fast development of web based computer that wishes to use them [3]. Web applications do applications. It offers development using CGI and WSGI. not incur publishing and distribution costs as opposed to Web development in Python is aided by the powerful traditional applications where the software (applications) frameworks such as Django, web2py, Pyramid, and Flask were published using CD’s and distributed. They need not be that Python supports. Thus Python promises to emerge as installed at each client host; rather they are placed at a one of the preferred choice language for web applications. central server and accessed by large number of clients. Since a web application is managed centrally, application updates KEYWORDS and data backups can be performed easily. The web Python, Web server application development, frameworks, applications are easily accessible irrespective of the WSGI, CGI, PHP, Ruby boundaries of space and time. -

Odoo Development Cookbook
Odoo Development Cookbook Build effective applications by applying Odoo development best practices Holger Brunn Alexandre Fayolle Daniel Reis BIRMINGHAM - MUMBAI Odoo Development Cookbook Copyright © 2016 Packt Publishing All rights reserved. No part of this book may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, without the prior written permission of the publisher, except in the case of brief quotations embedded in critical articles or reviews. Every effort has been made in the preparation of this book to ensure the accuracy of the information presented. However, the information contained in this book is sold without warranty, either express or implied. Neither the authors, nor Packt Publishing, and its dealers and distributors will be held liable for any damages caused or alleged to be caused directly or indirectly by this book. Packt Publishing has endeavored to provide trademark information about all of the companies and products mentioned in this book by the appropriate use of capitals. However, Packt Publishing cannot guarantee the accuracy of this information. First published: April 2016 Production reference: 1260416 Published by Packt Publishing Ltd. Livery Place 35 Livery Street Birmingham B3 2PB, UK. ISBN 978-1-78588-364-4 www.packtpub.com FM-2 Credits Authors Project Coordinator Holger Brunn Kinjal Bari Alexandre Fayolle Daniel Reis Proofreader Safis Editing Reviewers Guewen Baconnier Indexer Monica Ajmera Mehta Stefan Rijnhart Production Coordinator Acquisition Editor Arvindkumar Gupta Manish Nainani Cover Work Content Development Editor Arvindkumar Gupta Mehvash Fatima Technical Editors Menza Mathew Deepti Tuscano Copy Editors Merilyn Pereira Alpha Singh FM-3 About the Authors Holger Brunn has been a fervent open source advocate since he came in to contact with the open source market sometime in the nineties.