Cantharellus Formosus) Genet Size Using Co-Dominant Microsatellite Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Mycological Society of San Francisco • Jan. 2016, Vol. 67:05
The Mycological Society of San Francisco • Jan. 2016, vol. 67:05 Table of Contents JANUARY 19 General Meeting Speaker Mushroom of the Month by K. Litchfield 1 President Post by B. Wenck-Reilly 2 Robert Dale Rogers Schizophyllum by D. Arora & W. So 4 Culinary Corner by H. Lunan 5 Hospitality by E. Multhaup 5 Holiday Dinner 2015 Report by E. Multhaup 6 Bizarre World of Fungi: 1965 by B. Sommer 7 Academic Quadrant by J. Shay 8 Announcements / Events 9 2015 Fungus Fair by J. Shay 10 David Arora’s talk by D. Tighe 11 Cultivation Quarters by K. Litchfield 12 Fungus Fair Species list by D. Nolan 13 Calendar 15 Mushroom of the Month: Chanterelle by Ken Litchfield Twenty-One Myths of Medicinal Mushrooms: Information on the use of medicinal mushrooms for This month’s profiled mushroom is the delectable Chan- preventive and therapeutic modalities has increased terelle, one of the most distinctive and easily recognized mush- on the internet in the past decade. Some is based on rooms in all its many colors and meaty forms. These golden, yellow, science and most on marketing. This talk will look white, rosy, scarlet, purple, blue, and black cornucopias of succu- at 21 common misconceptions, helping separate fact lent brawn belong to the genera Cantharellus, Craterellus, Gomphus, from fiction. Turbinellus, and Polyozellus. Rather than popping up quickly from quiescent primordial buttons that only need enough rain to expand About the speaker: the preformed babies, Robert Dale Rogers has been an herbalist for over forty these mushrooms re- years. He has a Bachelor of Science from the Univer- quire an extended period sity of Alberta, where he is an assistant clinical profes- of slower growth and sor in Family Medicine. -

December, 2008
The Mycological Society of San Francisco December 2008, vol. 59:09 Send submissions to MycoDigest: An Old Friend Mycena News! Gets a New Name Brian A. Perry or years, one of the most commonly collected and highly sought after Fedible mushroom species in California has gone without a proper scientific name. Although many of you may not have realized this, the oak woodland chanterelle we so commonly collect here in the Bay Area and other regions of California, has had a name based upon a European species loosely applied to it. Undoubtedly, some of you have heard professionals or other knowledgeable fungophiles proclaim that our oak chanterelle is not the same species as that found in the coniferous forests of the Pacific Northwest, and that someone ought to put a good name on that species. Well, finally, in a forthcoming issue of the Mycena News is seeking scientific journal “Economic Botany,” David Arora and co-author Susie Dunham content pertaining to any of rectify this situation, providing the chanterelle so common to our California oak the following topics. Please woodlands and mixed evergreen forests with a valid scientific name,Cantharellus consider submitting an article californicus sp. nov. to: [email protected]. -Photos of this season’s most As indicated by Arora and beautiful mushrooms Dunham (2008), the species name Cantharellus cibarius -Foray reports and experiences Fr., based on material in the field collected in France, has -Your favorite recipes been applied at one time or another to all of the golden -Mushroom related poems and chanterelles we encounter artwork in California (excluding of course, the white chanterelle C. -

Toxic Fungi of Western North America
Toxic Fungi of Western North America by Thomas J. Duffy, MD Published by MykoWeb (www.mykoweb.com) March, 2008 (Web) August, 2008 (PDF) 2 Toxic Fungi of Western North America Copyright © 2008 by Thomas J. Duffy & Michael G. Wood Toxic Fungi of Western North America 3 Contents Introductory Material ........................................................................................... 7 Dedication ............................................................................................................... 7 Preface .................................................................................................................... 7 Acknowledgements ................................................................................................. 7 An Introduction to Mushrooms & Mushroom Poisoning .............................. 9 Introduction and collection of specimens .............................................................. 9 General overview of mushroom poisonings ......................................................... 10 Ecology and general anatomy of fungi ................................................................ 11 Description and habitat of Amanita phalloides and Amanita ocreata .............. 14 History of Amanita ocreata and Amanita phalloides in the West ..................... 18 The classical history of Amanita phalloides and related species ....................... 20 Mushroom poisoning case registry ...................................................................... 21 “Look-Alike” mushrooms ..................................................................................... -

Polyozellus Multiplex-An Example of Our Mycological Ignorance. Osprey 37:20-22
20 21 Polyozellus Multiplex - An Example of Our Mycological Ignorance by Andrus Voitk Polyozellus multiplex, the blue (or black) chanterelle, shown in Figure 1, grows as a large cluster of black or very dark bluish-purple fan- shaped leaves, arising out of a wide, common stalk. The sporulating surface, on the outside of each fan, is made up of folds, somewhat resembling thick, forked, decurrent gills. The spore print is white, giving mature specimens a dull bluish-gray appearance on the outside, Figure 1 - Polyozellus multiplex, Stanleyville Trail, contrasting with the shinier blackish purple on Gros Morne National Park, September, 2000. the inside of each leaf. The same trees are still there but this gorgeous ectomycorrhizal mushroom has not been seen in that Classical morphological taxonomy considered location since. I am now aware of three places where Polyozellus to be a relative of the chantarelle with this mushroom has been found to fruit. In 1-5 follow- its fold-like gills, along with the genus Gomphus, up years it has not been found to fruit in any of these whose sporulating surface is virtually identical to places again. that of P. multiplex. Recent DNA studies have left Gomphus as a distant relative of the chanterelle but placed P. multiplex with earth fans, Class Telephorales, Family Telephuraceae. It doesn’t seem to fit readily with either and no doubt the future has more family unrest in store for poor P. multiplex. Among interesting properties of this good edible* is its ability to slow certain cell replication. Animal experiments suggest P. multiplex may, among other things, prevent stomach cancer (1). -

Molecular Phylogeny and Morphology Reveal Three New Species of Cantharellus Within 20 M of One Another in Western Wisconsin, USA
Mycologia, 105(2), 2013, pp. 447–461. DOI: 10.3852/12-181 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 Molecular phylogeny and morphology reveal three new species of Cantharellus within 20 m of one another in western Wisconsin, USA Matthew J. Foltz morphological and DNA data have shown that C. Kathryn E. Perez1 cibarius in the United States is a species complex that Thomas J. Volk requires further taxonomic attention (Feibelman et Department of Biology, University of Wisconsin-La al. 1994, Dunham et al. 2003, Moncalvo et al. 2006, Crosse, 1725 State Street, La Crosse, Wisconsin 54601 Arora and Dunham 2008, Buyck and Hofstetter 2011). In this study we continue efforts to document North American Cantharellus diversity. Abstract: Three new species, Cantharellus phasmatis, In the past 50 years, several advancements in Cantharellus flavus and Cantharellus spectaculus, all taxonomy of Cantharellus have been made from previously considered Cantharellus cibarius, are de- morphological data. Smith (1968) described C. scribed in this study. The circumscription of these cibarius var. cibarius from Michigan, which he three species from C. cibarius and other Cantharellus believed to be the same as C. cibarius from Europe. species is supported by morphological differences Some of the key diagnostic features from this and nuclear DNA sequence data (nLSU, ITS, TEF1). description are the ‘‘egg-yellow or paler’’ hymenium, All were found under Quercus spp. in a small plot in the ‘‘pale-ochraceous’’ spore print and the incurved- Hixon Forest Park in La Crosse, Wisconsin, empha- margin becoming plane-to-wavy and finally broadly sizing the need for further taxonomic study of even infundibuliform. -

Il/Ill Signature Redactsignature Redacted for Privacy
AN ABSTRACT OF THE THESIS OF Matthew James Trappe for the degree of Master of Science in Forest Science presented on September 13, 2001. Title: Ecology of Craterellus tubaeformj',AI in Westerrj..Qron.il/Ill Signature redactSignature redacted for privacy. Abstract approved: A. Castellano Robert P. Griffith Craterellus tuba eformis is a small to medium-sized forest mushroom that is fairly common in the Douglas-fir/western hemlock forests of the Pacific Northwestern United States and is most often associated with decayed coarse woody debris. In this study, the mycorrhizae of Craterellus tubaeformis in western Oregon is identified by DNA analysis using polymerase chain reaction (PCR) amplification and restriction fragment length polymorphism (RFLP) typing, and the mantle morphology isdescribed. Host associations with western hemlock, Douglas-fir5 and Sitka spruce are identified using the same molecular techniques, with Craterellus tubaeformis most commonly associated with western hemlock. Differences in genetic sequences and host associations between western North America, eastern North America, and. Europe are presented, and the possibility that variants of Craterellus. tubaeformis from the different geographies might deserve their own species epithets is discussed. The dependency of Craterellus tubaeformis on late seral stands and abundance of coarse woody debris was quantified by surveying 64 plots in the Coast and Cascade ranges of western Oregon.Logistic regression showed that the odds of Craterellus tubaeformis occurrence increased with stand age and coarse woody debris (CWD) volumes, however it is often found in younger stands. The likelihood of Craterellus tubaeformis occurrence in a stand was highly correlated to the presence of western hemlock. Linear regression analysis showed no significant relationships between stand age, CWD volume, slope, elevation, or aspect on Craterellus tuba eformis biomass productivity, though well- decayed CWD was the substrate for 88% of the collected biomass. -

Bulletin of the Puget Sound Mycological Society No
BULLETIN OF THE PUGET SOUND MYCOLOGICAL SOCIETY Number 416 November 2005 MONSTER MOLD THREATENS HEALTH IN THE Clothes can be washed or dry cleaned, but most furniture is a SOUTH Julia Silverman and Marilynn Marchione loss. Ditto for carpeting, insulation, wallpaper, and drywall, which The Sporeprint, L.A. Myco. Soc., October 2005 no longer lives up to its name. Mattresses that didn’t get wet prob- ably have mold if they were in a room that did. “Anything with a New Orleans (AP) - Wearing goggles, gloves, ga- cushion you can forget about,” May said. loshes, and a mask, Veronica Randazzo lasted only The general advice is the same as when food is suspected of be- 10 minutes inside her home in St. Bernard Parish. Her eyes burned, her mouth filled with a salty taste, ing spoiled—when in doubt, throw it out. and she felt nauseous. Her 26-year-old daughter, When is professional help needed? “It’s simply a matter of ex- Alicia, also covered in gear, came out coughing. “That mold,” tent. If you’ve got small areas of mold, just a few square feet, it’s she said. “It smells like death.” something a homeowner can clean with 10 percent bleach,” said Anu Dixit, a fungus expert at Saint Louis University. Standing water after Hurricane Katrina created ideal growth condi- tions for mold and allowed it to penetrate so deep that experts fear Dixit studied mold after the Mississippi River floods in 1993 and that even studs of many homes are saturated and unsalvageable. 1994, and found cleaning measures often were ineffective, mainly Mold now forms an interior version of kudzu in the soggy South, because people started rebuilding too soon, before the surround- posing health dangers that will make many homes tear-downs and ing area was completely dry. -

Unece Standard Ffv-55 Chanterelles
UNECE STANDARD FFV-55 concerning the marketing and commercial quality control of CHANTERELLES 2017 EDITION UNITED NATIONS New York and Geneva, 2017 FFV-55: Chanterelles – 2017 NOTE Working Party on Agricultural Quality Standards The commercial quality standards developed by the UNECE Working Party on Agricultural Quality Standards help facilitate international trade, encourage high-quality production, improve profitability and protect consumer interests. UNECE standards are used by governments, producers, traders, importers and exporters, and other international organizations. They cover a wide range of agricultural products, including fresh fruit and vegetables, dry and dried produce, seed potatoes, meat, cut flowers, eggs and egg products. Any member of the United Nations can participate, on an equal footing, in the activities of the Working Party. For more information on agricultural standards, please visit our website <www.unece.org/trade/agr>. The present revised Standard for Chanterelles is based on document ECE/TRADE/C/WP.7/2014/11, reviewed and adopted by the Working Party at its seventieth session. Aligned with the Standard Layout (2017) The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the United Nations Secretariat concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Mention of company names or commercial products does not imply -

Cesium Radioisotope Content of Wild Edible Fungi, Mineral Soil, and Surface Litter in Western North America After the Fukushima
1 Cesium radioisotope content of wild edible fungi, mineral soil, and surface litter in western 2 North America after the Fukushima nuclear accident. 3 4 Matthew J. Trappea, Leah D. Mincb, Kimberly S. Kittredgec, Jeremias W. Pinkd 5 6 a Department of Forest Ecosystems and Society, 321 Richardson Hall, Oregon State University, 7 Corvallis, Oregon, USA, 97331, [email protected], corresponding author, Tel. 01- 8 541-737-6072 9 10 b Radiation Center, 100 Radiation Center, Oregon State University, Corvallis, Oregon, USA, 11 97331, [email protected] 12 13 c Northwest Mycological Consultants, 702 NW 4th Street, Corvallis, Oregon, USA, 97330, 14 [email protected] 15 16 d Department of Anthropology, 238 Waldo Hall, Oregon State University, Corvallis, Oregon, 17 USA, 97331, [email protected] 1 18 ABSTRACT 19 We measured activity levels of cesium radioisotopes 134Cs and 137Cs in wild edible fungi, 20 mineral soil, and surface litter of the west coast of North America from southern California to 21 northern Vancouver Island after the Fukushima nuclear accident. All activity measurements 22 were below governmental limits for human health. 137Cs activity increased to the north in 23 mineral soils and fungal samples, while 134Cs activity increased to the south in surface litter 24 samples. Chanterelles did not significantly bioconcentrate either radioisotope, but chanterelle 25 activity levels were correlated with those of mineral soil. Activity levels demonstrated a high 26 degree of variability, even in samples from the same site. In most cases the level of 137Cs 27 activity was substantially higher than that of 134Cs, suggesting that 137Cs was present in the 28 environment prior to the Fukushima release. -

Complete References List
Aanen, D. K. & T. W. Kuyper (1999). Intercompatibility tests in the Hebeloma crustuliniforme complex in northwestern Europe. Mycologia 91: 783-795. Aanen, D. K., T. W. Kuyper, T. Boekhout & R. F. Hoekstra (2000). Phylogenetic relationships in the genus Hebeloma based on ITS1 and 2 sequences, with special emphasis on the Hebeloma crustuliniforme complex. Mycologia 92: 269-281. Aanen, D. K. & T. W. Kuyper (2004). A comparison of the application of a biological and phenetic species concept in the Hebeloma crustuliniforme complex within a phylogenetic framework. Persoonia 18: 285-316. Abbott, S. O. & Currah, R. S. (1997). The Helvellaceae: Systematic revision and occurrence in northern and northwestern North America. Mycotaxon 62: 1-125. Abesha, E., G. Caetano-Anollés & K. Høiland (2003). Population genetics and spatial structure of the fairy ring fungus Marasmius oreades in a Norwegian sand dune ecosystem. Mycologia 95: 1021-1031. Abraham, S. P. & A. R. Loeblich III (1995). Gymnopilus palmicola a lignicolous Basidiomycete, growing on the adventitious roots of the palm sabal palmetto in Texas. Principes 39: 84-88. Abrar, S., S. Swapna & M. Krishnappa (2012). Development and morphology of Lysurus cruciatus--an addition to the Indian mycobiota. Mycotaxon 122: 217-282. Accioly, T., R. H. S. F. Cruz, N. M. Assis, N. K. Ishikawa, K. Hosaka, M. P. Martín & I. G. Baseia (2018). Amazonian bird's nest fungi (Basidiomycota): Current knowledge and novelties on Cyathus species. Mycoscience 59: 331-342. Acharya, K., P. Pradhan, N. Chakraborty, A. K. Dutta, S. Saha, S. Sarkar & S. Giri (2010). Two species of Lysurus Fr.: addition to the macrofungi of West Bengal. -
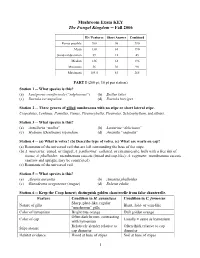
Mushroom Exam KEY the Fungal Kingdom -- Fall 2006
Mushroom Exam KEY The Fungal Kingdom -- Fall 2006 ID / Features Short Answer Combined Points possible 200 90 290 Mean 130 61 190 Standard deviation 39 12 45 Median 136 61 196 Minimum 36 36 90 Maximum 184.5 83 265 PART I (200 pt; 10 pt per station) Station 1 -- What species is this? (a) Laetiporus conifericola (“sulphureus”) (b) Suillus lakei (c) Russula xerampelina (d) Russula brevipes Station 2 -- Three genera of gilled mushrooms with no stipe or short lateral stipe. Crepidotus, Lentinus, Panellus, Panus, Pleurocybella, Pleurotus, Schizophyllum, and others. Station 3 -- What species is this? (a) Armillaria “mellea” (b) Lactarius “deliciosus” (c) Hydnum (Dentinum) repandum (d) Amanita “vaginata” Station 4 -- (a) What is volva? (b) Describe type of volva. (c) What are warts on cap? (a) Remnants of the universal veil that are left surrounding the base of the stipe. (b) A. muscaria: zoned, or ringed; A. pantherina: collared, or circumsessile, bulb with a free rim of tissue; A. phalloides: membranous saccate (broad and cup-like); A. vaginata: membranous saccate (narrow and upright, may be constricted) (c) Remnants of the universal veil. Station 5 -- What species is this? (a) Aleuria aurantia (b) Amanita phalloides (c) Ganoderma oregonense (tsugae) (d) Boletus edulis Station 6 -- Keep the Coop honest; distinguish golden chanterelle from false chanterelle. Feature Condition in H. aurantiaca Condition in C. formosus Sharp, plate-like, regular Nature of gills Blunt, fold- or vein-like “mushroom” gills Color of hymenium Bright true orange Dull golden orange Often dark brown, contrasting Color of cap Usually ± same as hymenium with hymenium Relatively slender relative to Often thick relative to cap Stipe stature cap diameter diameter Habitat evidence Wood at base of stipes Soil at base of stipes 1 Feature Condition in H. -

MYCENA NEWS VOL 69:02 Submissions for the January Newsletter Are Due by December 20Th
Mycological Society of San Francisco DEC 2017 MYCENA NEWS VOL 69:02 Submissions for the January newsletter are due by December 20th TABLE OF CONTENTS Holiday Dinner Eric Multhaup 1 Ethnomyco Culture Cusp Rivkah Khanin 10 Fungus Fair Madhu Kottalam 1 Dyer’s Niche Michael Spurgeon 11 President’s Post Tyler Taunton 3 Cultivation Quadrant Ken Litchfield 12 Hospitality Eric Multhaup 3 Calendar 12 Culinary Corner Morgan Evans 4 Mushroom Sightings 13 Mendo Camp Species List Fred Stevens 7 Mycena News Submissions 14 Holiday Dinner Fungus Fair Eric Multhaup Madhu Kottalam DECEMBER 11TH SIGN UP NOW You are invited to sign up NOW on the website for the December 11th Holiday Dinner, set-up at 6:00 p.m., appetizers at 7:00, dinner to follow, and seasonal jollity available at all times. Registration ends on December 7, 2017 Ticket prices for general members are $40, and $45 for guests. Ticket prices for student members, or members on fixed incomes are $20, and $25 for guests. Please pay by PayPal if you can; other- wise send a check to Eric Multhaup at 222 Julia Avenue, Mill Valley, CA 94941. We need volunteers for set-up, check-in, and 48th Annual Fungus Fair 2017 clean-up as usual. Please e-mail Eric at mullew@ Join us Sunday, December 3rd, for the 48th Annual comcast.net if you can volunteer. Fungus Fair—A celebration of fungi featuring wild mushroom displays, lectures, cooking demonstra- Location: Hall of Flowers—Golden Gate Park tions, activities, and vendors. 1199 9th Ave. at Lincoln Way We’ll have more than a dozen taxonomically San Francisco, CA 94122 arranged mushroom display tables to better learn their familial relationships, multitudes of specialty continued p.