Characterisation and Cross-Amplification of Polymorphic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Local Nature Reserve Management Plan 2020 – 2024
Bisley Road Cemetery, Stroud Local Nature Reserve Management Plan 2020 – 2024 Prepared for Stroud Town Council CONTENTS 1 VISION STATEMENT 2 POLICY STATEMENTS 3 GENERAL DESCRIPTION 3.1 General background information 3.1.1 Location and site boundaries Map 1 Site Location 3.1.2 Tenure Map 2 Schedule Plan 3.1.3 Management/organisational infrastructure 3.1.4 Site infrastructure 3.1.5 Map coverage 3.2 Environmental information 3.2.1 Physical 3.2.2 Biological 3.2.2.1 Habitats Map 3 Compartment Map – Old Cemetery Map 4 Compartment Map – New Cemetery 3.2.2.2 Flora 3.2.2.3 Fauna 3.3 Cultural 3.3.1 Past land use 3.3.2 Present land use 3.3.3 Past management for nature conservation 3.3.4 Present legal status 4 NATURE CONSERVATION FEATURES OF INTEREST 4.1 Identification and confirmation of conservation features 4.2 Objectives 4.2.1 Unimproved grassland 4.2.1.1 Summary description 4.2.1.2 Management objectives 4.2.1.3 Performance indicators 4.2.1.4 Conservation status 4.2.1.5 Rationale 4.2.1.6 Management projects 4.2.2 Trees and Woodland 4.2.2.1 Summary description 4.2.2.2 Management objectives 4.2.2.3 Performance indicators 4.2.2.4 Conservation status 4.2.2.5 Rationale 4.2.2.6 Management projects 4.2.3 Lichens 4.2.3.1 Summary description 4.2.3.2 Management objectives 4.2.3.3 Performance indicators 4.2.3.4 Conservation status 4.2.3.5 Rationale 4.2.3.6 Management projects 4.3 Rationale & Proposals per compartment Bisley Rd Cemetery Mgmt Plan 2020-2024 2 5 HISTORIC INTEREST 5.1 Confirmation of conservation features 5.2 Objectives 5.3 Rationale 6 STAKEHOLDERS 6.1 Evaluation 6.2 Management projects 7 ACCESS / TOURISM 7.1 Evaluation 7.2 Management objectives 8 INTERPRETATION 8.1 Evaluation 8.2 Management Projects 9 OPERATIONAL OBJECTIVES 9.1 Operational objectives 9.2 Management projects 10 WORK PLAN Appendix 1 Species List Bisley Rd Cemetery Mgmt Plan 2020-2024 3 1 VISION STATEMENT Stroud Town Council are committed to conserving Stroud Cemetery to: • Enable the people of Stroud to always have a place of peace and quiet reflection and recreation. -

A Contribution to the Aphid Fauna of Greece
Bulletin of Insectology 60 (1): 31-38, 2007 ISSN 1721-8861 A contribution to the aphid fauna of Greece 1,5 2 1,6 3 John A. TSITSIPIS , Nikos I. KATIS , John T. MARGARITOPOULOS , Dionyssios P. LYKOURESSIS , 4 1,7 1 3 Apostolos D. AVGELIS , Ioanna GARGALIANOU , Kostas D. ZARPAS , Dionyssios Ch. PERDIKIS , 2 Aristides PAPAPANAYOTOU 1Laboratory of Entomology and Agricultural Zoology, Department of Agriculture Crop Production and Rural Environment, University of Thessaly, Nea Ionia, Magnesia, Greece 2Laboratory of Plant Pathology, Department of Agriculture, Aristotle University of Thessaloniki, Greece 3Laboratory of Agricultural Zoology and Entomology, Agricultural University of Athens, Greece 4Plant Virology Laboratory, Plant Protection Institute of Heraklion, National Agricultural Research Foundation (N.AG.RE.F.), Heraklion, Crete, Greece 5Present address: Amfikleia, Fthiotida, Greece 6Present address: Institute of Technology and Management of Agricultural Ecosystems, Center for Research and Technology, Technology Park of Thessaly, Volos, Magnesia, Greece 7Present address: Department of Biology-Biotechnology, University of Thessaly, Larissa, Greece Abstract In the present study a list of the aphid species recorded in Greece is provided. The list includes records before 1992, which have been published in previous papers, as well as data from an almost ten-year survey using Rothamsted suction traps and Moericke traps. The recorded aphidofauna consisted of 301 species. The family Aphididae is represented by 13 subfamilies and 120 genera (300 species), while only one genus (1 species) belongs to Phylloxeridae. The aphid fauna is dominated by the subfamily Aphidi- nae (57.1 and 68.4 % of the total number of genera and species, respectively), especially the tribe Macrosiphini, and to a lesser extent the subfamily Eriosomatinae (12.6 and 8.3 % of the total number of genera and species, respectively). -

Folk Taxonomy, Nomenclature, Medicinal and Other Uses, Folklore, and Nature Conservation Viktor Ulicsni1* , Ingvar Svanberg2 and Zsolt Molnár3
Ulicsni et al. Journal of Ethnobiology and Ethnomedicine (2016) 12:47 DOI 10.1186/s13002-016-0118-7 RESEARCH Open Access Folk knowledge of invertebrates in Central Europe - folk taxonomy, nomenclature, medicinal and other uses, folklore, and nature conservation Viktor Ulicsni1* , Ingvar Svanberg2 and Zsolt Molnár3 Abstract Background: There is scarce information about European folk knowledge of wild invertebrate fauna. We have documented such folk knowledge in three regions, in Romania, Slovakia and Croatia. We provide a list of folk taxa, and discuss folk biological classification and nomenclature, salient features, uses, related proverbs and sayings, and conservation. Methods: We collected data among Hungarian-speaking people practising small-scale, traditional agriculture. We studied “all” invertebrate species (species groups) potentially occurring in the vicinity of the settlements. We used photos, held semi-structured interviews, and conducted picture sorting. Results: We documented 208 invertebrate folk taxa. Many species were known which have, to our knowledge, no economic significance. 36 % of the species were known to at least half of the informants. Knowledge reliability was high, although informants were sometimes prone to exaggeration. 93 % of folk taxa had their own individual names, and 90 % of the taxa were embedded in the folk taxonomy. Twenty four species were of direct use to humans (4 medicinal, 5 consumed, 11 as bait, 2 as playthings). Completely new was the discovery that the honey stomachs of black-coloured carpenter bees (Xylocopa violacea, X. valga)were consumed. 30 taxa were associated with a proverb or used for weather forecasting, or predicting harvests. Conscious ideas about conserving invertebrates only occurred with a few taxa, but informants would generally refrain from harming firebugs (Pyrrhocoris apterus), field crickets (Gryllus campestris) and most butterflies. -

The Role of Ant Nests in European Ground Squirrel's (Spermophilus
Biodiversity Data Journal 7: e38292 doi: 10.3897/BDJ.7.e38292 Short Communications The role of ant nests in European ground squirrel’s (Spermophilus citellus) post-reintroduction adaptation in two Bulgarian mountains Maria Kachamakova‡‡, Vera Antonova , Yordan Koshev‡ ‡ Institute of Biodiversity and Ecosystem Research at Bulgarian Academy of Sciences, Sofia, Bulgaria Corresponding author: Maria Kachamakova ([email protected]) Academic editor: Ricardo Moratelli Received: 16 Jul 2019 | Accepted: 09 Sep 2019 | Published: 07 Oct 2019 Citation: Kachamakova M, Antonova V, Koshev Y (2019) The role of ant nests in European ground squirrel’s (Spermophilus citellus) post-reintroduction adaptation in two Bulgarian mountains. Biodiversity Data Journal 7: e38292. https://doi.org/10.3897/BDJ.7.e38292 Abstract The European ground squirrel (Spermophilus citellus) is a vulnerable species, whose populations are declining throughout its entire range in Central and South-Eastern Europe. To a great extent, its conservation depends on habitat restoration, maintenance and protection. In order to improve the conservation status of the species, reintroductions are increasingly applied. Therefore, researchers focus their attention on factors that facilitate these activities and contribute to their success. In addition to the well-known factors like grass height and exposition, others, related to the underground characteristics, are more difficult to evaluate. The presence of other digging species could help this evaluation. Here, we present two reintroduced ground squirrel colonies, where the vast majority of the burrows are located in the base of anthills, mainly of yellow meadow ant (Lasius flavus). This interspecies relationship offers numerous advantages for the ground squirrel and is mostly neutral for the ants. -

Reproduction and Dispersal in an Antassociated Root Aphid Community
Molecular Ecology (2012) doi: 10.1111/j.1365-294X.2012.05701.x Reproduction and dispersal in an ant-associated root aphid community A. B. F. IVENS,*† D. J. C. KRONAUER,†‡ I. PEN,*F.J.WEISSING*andJ.J.BOOMSMA† *Theoretical Biology, Centre for Ecological and Evolutionary Studies, University of Groningen, Nijenborgh 7, 9747 AG Groningen, The Netherlands, †Centre for Social Evolution, Department of Biology, University of Copenhagen, Universitetsparken 15, DK-2100 Copenhagen, Denmark, ‡Laboratory of Insect Social Evolution, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA Abstract Clonal organisms with occasional sex are important for our general understanding of the costs and benefits that maintain sexual reproduction. Cyclically parthenogenetic aphids are highly variable in their frequency of sexual reproduction. However, studies have mostly focused on free-living aphids above ground, whereas dispersal constraints and dependence on ant-tending may differentially affect the costs and benefits of sex in subterranean aphids. Here, we studied reproductive mode and dispersal in a community of root aphids that are obligately associated with the ant Lasius flavus. We assessed the genetic population structure of four species (Geoica utricularia, Tetraneura ulmi, Forda marginata and Forda formicaria) in a Dutch population and found that all species reproduce predominantly if not exclusively asexually, so that populations consist of multiple clonal lineages. We show that population viscosity is high and winged aphids rare, consistent with infrequent horizontal transmission between ant host colonies. The absence of the primary host shrub (Pistacia) may explain the absence of sex in three of the studied species, but elm trees (Ulmus) that are primary hosts of the fourth species (T. -
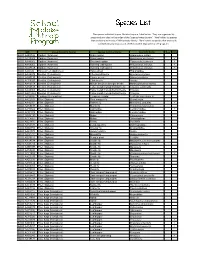
Species List
The species collected in your Malaise trap are listed below. They are organized by group and are listed in the order of the 'Species Image Library'. ‘New’ refers to species that are brand new to our DNA barcode library. 'Rare' refers to species that were only collected in your trap out of all 67 that were deployed for the program. BIN Group (Scientific Name) Species Common Name Scientific Name New Rare BOLD:AAB5726 Spiders (Araneae) Grass spider Agelenopsis potteri BOLD:AAD6926 Spiders (Araneae) Ghost spider Anyphaena pectorosa BOLD:AAH0002 Spiders (Araneae) Sheetweb spider Tapinocyba hortensis BOLD:AAN9005 Spiders (Araneae) Running crab spider Philodromus minutus BOLD:ACX7149 Spiders (Araneae) Running crab spider Philodromus minutus BOLD:AAI4346 Harvestmen (Opiliones) Harvestman Phalangiidae BOLD:AAU6970 Beetles (Coleoptera) Checkered beetle Enoclerus nigripes BOLD:AAH0130 Beetles (Coleoptera) Clover weevil Sitona hispidulus BOLD:ACP9027 Beetles (Coleoptera) Click beetle Aeolus BOLD:AAN6154 Beetles (Coleoptera) Minute brown scavenger beetle Melanophthalma inermis BOLD:AAN6148 Beetles (Coleoptera) False metallic wood-boring beetle Trixagus carinicollis BOLD:AAU6966 Beetles (Coleoptera) False metallic wood-boring beetle Trixagus BOLD:ABW2869 Beetles (Coleoptera) False metallic wood-boring beetle Trixagus BOLD:AAG9897 Earwigs (Dermaptera) European earwig Forficula auricularia-A BOLD:AAG2513 Flies (Diptera) Root maggot fly Eustalomyia BOLD:AAH2351 Flies (Diptera) Robber fly Machimus sadyates BOLD:AAN5137 Flies (Diptera) March fly Penthetria -

Book of Abstracts Goettingen O
Imprint Geschäftstelle der Ge‐ Verhandlungen der Gesellschaft sellschaft für Ökologie für Ökologie, Band 45 Institut für Ökologie Herausgegeben im Auftrag der Gesell‐ Technische Universität schaft für Ökologie von apl. Prof. Dr. Berlin Rothenburgerstr. Michael Bredemeier, Sektion Waldöko‐ 12 systemforschung, Zentrum für Biodiver‐ D‐12165 Berlin sität und nachhaltige Landnutzung (CBL), Tel.: 030‐ Georg‐August‐Universität Göttingen 31471396 Fax.: 030‐31471355 © Gesellschaft für Ökologie Göttingen 2015, ISSN: 0171‐1113 Contact: KCS Kuhlmann Convention Service Universität Göttingen Heike Kuhmann Prof. Dr. Michael Bredemeier, Rue des Chênes Sektion Waldökosystemfor‐ 12 CH‐2800 De‐ schung, Büsgenweg 2 lémont D‐ 37077 Göttingen + 41‐32‐4234384 info@kcs‐convention.com The 45th annual conference of the Ecological Society of Germany, Austria and Switzer‐ land (GfÖ) is taking place from 31st August to 04th September 2015 at the Universi‐ ty of Göttingen. Host of the conference is the Centre for Biodiversity and Sustainable Land Use (CBL), University of Göttingen. Local Organizing Committee Chair: Prof. Dr. Christian Ammer, University of Göt‐ tingen Co‐Chairs: Prof. Dr. Teja Tscharntke, Prof. Dr. Kerstin Wie‐ gand, University of Göttingen Proceedings editors: Prof. Dr. Michael Bredemeier, Dr. Juliane Steckel, Ursula Seele, University of Göttingen Field Trip planning: Dr. Simone Pfeiffer, University of Göttingen Scientific Programm Committee Prof. Dr. Christian Ammer, Prof. Dr. Michael Bredemeier, Prof. Dr. Alexander Knohl, Prof. Dr. Holger Kreft, Dr. Simone Pfeiffer, PD Dr. Martin Potthoff, Prof. Dr. Stefan Scheu, Prof. Dr. Teja Tscharntke Dr. G. Wiedey, Prof. Dr. Kerstin Wiegand The book is also available for download as electronic document on the conference web site (www. gfoe‐2015.de) Production: Centre for Biodiversity and Sustainable Land Use (CBL), University of Göttingen, Büsgenweg 2, 37077 Göttingen, Germany Editors: Prof. -

Irish Ants (Hymenoptera, Formicidae): Distribution, Conservation and Functional Relationships
Irish Ants (Hymenoptera, Formicidae): Distribution, Conservation and Functional Relationships Submitted by: Dipl. Biol. Robin Niechoj Supervisor: Prof. John Breen Submitted in accordance with the academic requirements for the Degree of Doctor of Philosophy to the Department of Life Sciences, Faculty of Science and Engineering, University of Limerick Limerick, April 2011 Declaration I hereby declare that I am the sole author of this thesis and that it has not been submitted for any other academic award. References and acknowledgements have been made, where necessary, to the work of others. Signature: Date: Robin Niechoj Department of Life Sciences Faculty of Science and Engineering University of Limerick ii Acknowledgements/Danksagung I wish to thank: Dr. John Breen for his supervision, encouragement and patience throughout the past 5 years. His infectious positive attitude towards both work and life was and always will be appreciated. Dr. Kenneth Byrne and Dr. Mogens Nielsen for accepting to examine this thesis, all the CréBeo team for advice, corrections of the report and Dr. Olaf Schmidt (also) for verification of the earthworm identification, Dr. Siobhán Jordan and her team for elemental analyses, Maria Long and Emma Glanville (NPWS) for advice, Catherine Elder for all her support, including fieldwork and proof reading, Dr. Patricia O’Flaherty and John O’Donovan for help with the proof reading, Robert Hutchinson for his help with the freeze-drying, and last but not least all the staff and postgraduate students of the Department of Life Sciences for their contribution to my work. Ich möchte mich bedanken bei: Katrin Wagner für ihre Hilfe im Labor, sowie ihre Worte der Motivation. -

Aspects of the Biology and Taxonomy of British Myrmecophilous Root Aphids
Aspects of the biology and taxonomy of British myrmecophilous root aphids. by Richard George PAUL, BoSc.(Lond.),17.90.(Glcsgow). Thesis submitted for the Degree of Doctor of Philosopk,. of the University of London and for the Diploma of Imperial Colleao Decemi)or 1977. Dopartaent o:vv. .;:iotory)c Cromwell Road. 1.0;AMOH )t=d41/: -1- ABSTRACT. This thesis concerns the biology and taxonomy of root feeding aphids associated with British ants. A root aphid for the purposes of this thesis is defined as an aphid which, during at least part of its life cycle feeds either (a) beneath the normal soil surface or (b) beneath a tent of soil that has been placed over it by ants. The taxonomy of the genera Paranoecia and Anoecia has been revised and some synonomies proposed. Chromosome numbers have been discovered for Anoecia spp. and are used to clarify the taxonomy. The biology of Anoecia species has been studied and new facts about their life cycles have been discovered. A key is given to the British r European, African and North American species of Anoeciinae and this is included in a key to British myrmecophilous root aphids. Suction trap catches (1968-1976) from about twenty British traps have been used as a record of seasonal flight patterns for root aphids. All the Anoecia species caught in 1975 and 1976 were identified on the basis of new taxonomic work. Catches which had formerly all been identified as Anoecia corni were found to be A. corni, A. varans and A. furcata. The information derived from the catches was used to plot relative abundance and distribution maps for the three species. -

Aphids (Hemiptera: Aphidoidea) Associated with Native Trees in Malta (Central Mediterranean)
BULLETIN OF THE ENTOMOLOGICAL SOCIETY OF MALTA (2009) Vol. 2 : 81-93 Aphids (Hemiptera: Aphidoidea) associated with native trees in Malta (Central Mediterranean) David MIFSUD1, Nicolás PÉREZ HIDALGO2 & Sebastiano BARBAGALLO3 ABSTRACT. In the present study 25 aphid species which are known to be associated with trees in the Maltese Islands are recorded. Of these, 18 species represent new records; these include Aphis craccivora, Brachyunguis tamaricis, Cavariella aegopodii, Chaitophorus capreae, C. populialbae, Cinara cupressi, C. maghrebica, C. palaestinensis, Essigella californica, Eulachnus rileyi, E. tuberculostemmatus, Hoplocallis picta, Lachnus roboris, Myzocallis schreiberi, Tetraneura nigriabdominalis, Thelaxes suberi, Tinocallis takachihoensis and Tuberolachnus salignus. A number of the above mentioned species alternate hosts between the primary host, being the tree species, and secondary hosts being mainly roots of grasses. The record of Tetraneura ulmi could be incorrect and could possibly be referred to T. nigriabdominalis. Most of the aphid species recorded in the present study have restricted distribution in the Maltese Islands due to the rarity of their host trees. This is particularly so for those aphids associated with Populus, Quercus, Salix and Ulmus whose conservation should be addressed. INTRODUCTION Aphids belong to the suborder Sternorrhyncha within the order Hemiptera, along with scale insects, jumping plant-lice, or psylloids, and whiteflies. The Aphidoidea is predominantly a northern temperate group, richest in species in North America, Europe, and Central and Eastern Asia. A general feature of the life cycle of aphids is their parthenogenetic generations exploiting active growing plant parts and a sexual generation resulting in an overwintering diapause egg. The known world fauna of aphids consists of approximately 4400 described species placed in nearly 500 currently accepted genera. -

Scientific Name Common Name Taxon Group Aceria Pseudoplatani
Scientific Name Common Name Taxon Group Aceria pseudoplatani Aceria pseudoplatani acarine (Acari) Alabidocarpus acarine (Acari) Phytoptus avellanae Phytoptus avellanae acarine (Acari) Tetranychidae Tetranychidae acarine (Acari) Tetranychus urticae Tetranychus urticae acarine (Acari) Trombidiidae Trombidiidae acarine (Acari) Bufo bufo Common Toad amphibian Lissotriton helveticus Palmate Newt amphibian Lissotriton vulgaris Smooth Newt amphibian Rana Frog amphibian Rana temporaria Common Frog amphibian Triturus Newt amphibian Triturus cristatus Great Crested Newt amphibian Triturus helveticus Palmate Newt amphibian Annelida Worms annelid Eisenia fetida Manure Worm annelid Erpobdella testacea Erpobdella testacea annelid Lumbricus terrestris Common Earthworm annelid Acanthis flammea/cabaret agg. Redpoll (Common\Lesser) agg. bird Accipiter nisus Sparrowhawk bird Acrocephalus schoenobaenus Sedge Warbler bird Aegithalos caudatus Long-tailed Tit bird Alauda arvensis Skylark bird Anas platyrhynchos Mallard bird Anthus pratensis Meadow Pipit bird Apodidae Swifts bird Apus apus Swift bird Scientific Name Common Name Taxon Group Ardea cinerea Grey Heron bird Aythya fuligula Tufted Duck bird Branta canadensis Canada Goose bird Buteo buteo Buzzard bird Caprimulgus europaeus Nightjar bird Carduelis carduelis Goldfinch bird Certhia familiaris Treecreeper bird Chloris chloris Greenfinch bird Chroicocephalus ridibundus Black-headed Gull bird Cinclus cinclus Dipper bird Coloeus monedula Jackdaw bird Columba Pigeon bird Columba livia Feral Pigeon/Rock Dove -

Histological Study of Galls Induced by Aphids on Leaves of Ulmus Minor: Tetraneura Ulmi Induces Globose Galls and Eriosoma Ulmi Induces Pseudogalls
Arthropod-Plant Interactions (2013) 7:643–650 DOI 10.1007/s11829-013-9278-8 ORIGINAL PAPER Histological study of galls induced by aphids on leaves of Ulmus minor: Tetraneura ulmi induces globose galls and Eriosoma ulmi induces pseudogalls Rafael A´ lvarez • Silvia Gonza´lez-Sierra • Adoracio´n Candelas • Jean-Jacques Itzhak Martinez Received: 25 April 2013 / Accepted: 18 September 2013 / Published online: 27 September 2013 Ó Springer Science+Business Media Dordrecht 2013 Abstract The histological study of galls may provide Keywords Gall Á Pseudogall Á Tetraneura ulmi Á information on the evolution of the organisms that induce Eriosoma ulmi Á Ulmus minor Á Histological study them. The walls of two aphid-induced galls on leaves of Ulmus minor were therefore studied histologically: a glo- bose gall induced by Tetraneura ulmi and a pseudogall Introduction induced by Eriosoma ulmi. Galls are regarded as extended phenotypes of aphids, and therefore, they can be used as More than 15,000 species of organisms capable of inducing tools for phylogenetic studies. The walls of the galls the formation of galls are known, among which there are induced by T. ulmi are not reminiscent of the ungalled leaf viruses, bacteria, algae, fungi, protozoa, rotifers, nema- structure of U. minor in any area, showing both cellular todes, mites and insects (Felt 1940). hypertrophy and hyperplasia. Galls induced by E. ulmi Among the gall-forming organisms are several insect resemble the leaf structure of U. minor in certain aspects, families, including various aphid families. Approximately but in most aspects they do not, showing only cellular 10–20 % of the 4,700 known aphid species worldwide are hypertrophy.