Anti-H3r2me2(Asym) Antibody
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Four Enzymes Cooperate to Displace Histone H1 During the First Minute of Hormonal Gene Activation
Downloaded from genesdev.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation Guillermo Pablo Vicent,1,2 A. Silvina Nacht,1 Jofre Font-Mateu, Giancarlo Castellano, Laura Gaveglia, Cecilia Ballare´, and Miguel Beato Centre de Regulacio´ Geno` mica (CRG), Universitat Pompeu Fabra (UPF), E-08003 Barcelona, Spain Gene regulation by external signals requires access of transcription factors to DNA sequences of target genes, which is limited by the compaction of DNA in chromatin. Although we have gained insight into how core histones and their modifications influence this process, the role of linker histones remains unclear. Here we show that, within the first minute of progesterone action, a complex cooperation between different enzymes acting on chromatin mediates histone H1 displacement as a requisite for gene induction and cell proliferation. First, activated progesterone receptor (PR) recruits the chromatin remodeling complexes NURF and ASCOM (ASC-2 [activating signal cointegrator-2] complex) to hormone target genes. The trimethylation of histone H3 at Lys 4 by the MLL2/MLL3 subunits of ASCOM, enhanced by the hormone-induced displacement of the H3K4 demethylase KDM5B, stabilizes NURF binding. NURF facilitates the PR-mediated recruitment of Cdk2/CyclinA, which is required for histone H1 displacement. Cooperation of ATP-dependent remodeling, histone methylation, and kinase activation, followed by H1 displacement, is a prerequisite for the subsequent displacement of histone H2A/H2B catalyzed by PCAF and BAF. Chromatin immunoprecipitation (ChIP) and sequencing (ChIP-seq) and expression arrays show that H1 displacement is required for hormone induction of most hormone target genes, some of which are involved in cell proliferation. -

Inferring Causal Relationships Among Different Histone Modifications and Gene Expression
Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Inferring Causal Relationships among Different Histone Modifications and Gene Expression Hong Yu*, Shanshan Zhu*, Bing Zhou*, Huiling Xue and Jing-Dong J. Han Chinese Academy of Sciences Key Laboratory of Molecular Developmental Biology, Center for Molecular Systems Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Datun Road, Beijing, 100101, China * These authors contributed equally to this work. Correspondence should be addressed to J.-D.J.H ([email protected]). Running title: histone modification and gene expression network Keywords: histone modification, gene expression, Bayesian network, causal relationship, epigenetics 1 Downloaded from genome.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Histone modifications are major epigenetic factors regulating gene expression. They play important roles in maintaining stem cell pluripotency and in cancer pathogenesis. Different modifications may combine to form complex ‘histone codes’. Recent high throughput technologies, such as ‘ChIP-chip’ and ‘ChIP-seq’, have generated high resolution maps for many histone modifications on the human genome. Here we use these maps to build a Bayesian network to infer causal and combinatorial relationships among histone modifications and gene expression. A pilot network derived by the same method among polycomb group (PcG) genes and H3K27 trimethylation is accurately supported by current literature. Our unbiased network model among histone modifications is also well supported by cross validation results. It not only confirmed already known relationships, such as those of H3K27me3 to gene silencing, H3K4me3 to gene activation, and the effect of bivalent modification of both H3K4me3 and H3K27me3, but also identified many other relationships that may predict new epigenetic interactions important in epigenetic gene regulation. -
![[Dimethyl Arg17] Antibody NBP2-59162](https://docslib.b-cdn.net/cover/3309/dimethyl-arg17-antibody-nbp2-59162-743309.webp)
[Dimethyl Arg17] Antibody NBP2-59162
Product Datasheet Histone H3 [Dimethyl Arg17] Antibody NBP2-59162 Unit Size: 100 ul Store at -20C. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews, Research Tools and Images at: www.novusbio.com/NBP2-59162 Updated 5/13/2021 v.20.1 Earn rewards for product reviews and publications. Submit a publication at www.novusbio.com/publications Submit a review at www.novusbio.com/reviews/destination/NBP2-59162 Page 1 of 3 v.20.1 Updated 5/13/2021 NBP2-59162 Histone H3 [Dimethyl Arg17] Antibody Product Information Unit Size 100 ul Concentration Please see the vial label for concentration. If unlisted please contact technical services. Storage Store at -20C. Avoid freeze-thaw cycles. Clonality Polyclonal Preservative 0.05% Sodium Azide Isotype IgG Purity Whole Antiserum Buffer Serum Target Molecular Weight 15 kDa Product Description Host Rabbit Gene ID 126961 Gene Symbol H3C14 Species Human Immunogen The exact sequence of the immunogen to this Histone H3 [Dimethyl Arg17] antibody is proprietary. Product Application Details Applications Western Blot, Chromatin Immunoprecipitation, Dot Blot, ELISA Recommended Dilutions Western Blot 1:250, Chromatin Immunoprecipitation 10 - 15 uL/IP, ELISA 1:1000 - 1:3000, Dot Blot 1:20000 Page 2 of 3 v.20.1 Updated 5/13/2021 Images Western Blot: Histone H3 [Dimethyl Arg17] Antibody [NBP2-59162] - Histone extracts of HeLa cells (15 ug) were analysed by Western blot using the antibody against H3R17me2(asym) diluted 1:250 in TBS- Tween containing 5% skimmed milk. Observed molecular weight is ~17 kDa. Chromatin Immunoprecipitation: Histone H3 [Dimethyl Arg17] Antibody [NBP2-59162] - ChIP assays were performed using human osteosarcoma (U2OS) cells, the antibody against H3R17me2(asym) and optimized PCR primer sets for qPCR. -

Symmetric-Di-Methyl-Histone H3 (Arg8) Rabbit
Symmetric -Di-methyl-Histone H3 (Arg8) rabbit pAb ( H3R8me2(sy) pAb) www.ptm-biolab.com.cn Cat#: PTM-672 [email protected] Species: Rabbit 0571-2883 3567 Size: 100 μl Form: supplied in liquid form Recommend Species Swissprot Molecular Application Immunogen dilution Reactivity ID Weight H3R8me2(sy) WB, Dot, ELISA 1:2000 for WB H, M, R, Mk P68431 17 KDa -KLH **Species reactivity is determined by WB. Kept at -20oC after reconstituted. *** Anti-rabbit secondary antibodies must be used to detect this antibody. Source/Purification: This product is produced by immunizing rabbits with a synthetic di-methyl(sy) peptide corresponding to residues surrounding Arg8 of human histone H3. Antibodies are purified by protein A-conjugated agarose followed by di-methylated(sy) histone H3 (Arg8) peptide affinity chromatography. Recommended Applications: ELISA, Dot blot, Western blot. Recommended antibody dilution: WB: 1:2000 NOTE: For WB, incubate membrane with diluted antibody in 5% nonfat milk, 1 x TBS, 0.1% Tween- 20 for two hours at room temperature with gentle shaking. Prepare working dilution immediately before use. Use at an assay dependent concentration. Optimal dilutions/concentrations should be determined by the end user. Not yet tested in other applications Scientific Description: Proteins post-translational Modification (PTM) is an important reversible event controlling protein activity. The amino-terminal tails of core histones undergo multiple modifications in multiple sites, termed as “histone code” or “epigenetic code”. Lysine methylation or arginine methylation in core histones is a major determinant for the formation of active and inactive regions of the genome and therefore plays vital roles in multiple cellular events. -

BMC Genomics Biomed Central
BMC Genomics BioMed Central Research article Open Access Determination of enriched histone modifications in non-genic portions of the human genome Jeffrey A Rosenfeld1,2,3, Zhibin Wang4, Dustin E Schones4, Keji Zhao4, Rob DeSalle3 and Michael Q Zhang*1 Address: 1Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724 USA, 2Department of Biology, New York University, New York, NY USA, 3American Museum of Natural History, New York, NY USA and 4Laboratory of Molecular Immunology, National Heart, Lung and Blood Institute, NIH, Bethesda, MD USA Email: Jeffrey A Rosenfeld - [email protected]; Zhibin Wang - [email protected]; Dustin E Schones - [email protected]; Keji Zhao - [email protected]; Rob DeSalle - [email protected]; Michael Q Zhang* - [email protected] * Corresponding author Published: 31 March 2009 Received: 8 September 2008 Accepted: 31 March 2009 BMC Genomics 2009, 10:143 doi:10.1186/1471-2164-10-143 This article is available from: http://www.biomedcentral.com/1471-2164/10/143 © 2009 Rosenfeld et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP- seq) has recently been used to identify the modification patterns for the methylation and acetylation of many different histone tails in genes and enhancers. Results: We have extended the analysis of histone modifications to gene deserts, pericentromeres and subtelomeres. Using data from human CD4+ T cells, we have found that each of these non- genic regions has a particular profile of histone modifications that distinguish it from the other non- coding regions. -

Le G´Enome En Action
LEGENOME´ EN ACTION SEQUENC´ ¸ AGE HAUT DEBIT´ ET EPIG´ ENOMIQUE´ Epig´enomique´ ? IFT6299 H2014 ? UdeM ? Mikl´osCs}ur¨os Regulation´ d’expression la transcription d’une region´ de l’ADN necessite´ ? liaisons proteine-ADN´ (facteur de transcription et son site reconnu) ? accessibilite´ de la chromatine REVIEWS Identification of regions that control transcription An initial step in the analysis of any gene is the identifi- cation of larger regions that might harbour regulatory control elements. Several advances have facilitated the prediction of such regions in the absence of knowl- edge about the specific characteristics of individual cis- Chromatin regulatory elements. These tools broadly fall into two categories: promoter (transcription start site; TSS) and enhancer detection. The methods are influenced Distal TFBS by sequence conservation between ORTHOLOGOUS genes (PHYLOGENETIC FOOTPRINTING), nucleotide composition and the assessment of available transcript data. Functional regulatory regions that control transcrip- tion rates tend to be proximal to the initiation site(s) of transcription. Although there is some circularity in the Co-activator complex data-collection process (regulatory sequences are sought near TSSs and are therefore found most often in these regions), the current set of laboratory-annotated regula- tory sequences indicates that sequences near a TSS are Transcription more likely to contain functionally important regulatory initiation complex Transcription controls than those that are more distal. However, specifi- initiation cation of the position of a TSS can be difficult. This is fur- ther complicated by the growing number of genes that CRM Proximal TFBS selectively use alternative start sites in certain contexts. Underlying most algorithms for promoter prediction is a Figure 1 | Components of transcriptional regulation. -

Arsenite Exposure Inhibits Histone Acetyltransferase P300 For
!!!!!! "#$%&'(%!%)*+$,#%!'&-'.'($!-'$(+&%!"/%(01(#"&$2%#"$%!*344!2+#! "((%&,"('&5!-3678"/!"(!%&-"&/%#$!'&!1+9:;+$%!%)*+$%;!<+,$%! %<.#0+&'/!2'.#+.1"$(!/%11$! ! ! ! =>! ! 0?@!ABC! ! "!DEFFGHI?IEJ@!FC=KEIIGD!IJ!LJB@F!-JMNE@F!,@EOGHFEI>!E@!PJ@QJHKEI>!REIB!IBG! HGSCEHGKG@IF!QJH!IBG!DGTHGG!JQ!<?FIGH!JQ!$PEG@PG! ! .?UIEKJHGV!<?H>U?@D! ! "MHEUV!74WX! Y!0?@!ABC!74WX! ! "UU!#ETBIF!#GFGHOGD! ! ! !"#$%&'$( .JIB!GMEDGKEJUJTEP?U!E@OGFIET?IEJ@F!?@D!?@EK?U!FICDEGF!B?OG!UE@NGD!?HFG@EP! PJ@I?KE@?IGD!R?IGH!IJ!P?@PGHFZ!.GFEDGF!TG@JIJ[EPEI>V!?HFG@EP!G[MJFCHG:HGU?IGD! M?IBJTG@GFEF!JQ!DEFG?FG!EF!REDGU>!PJ@FEDGHGD!IBHJCTB!GMETG@GIEP!KGPB?@EFKF\! BJRGOGHV!IBG!C@DGHU>E@T!KGPB?@EFK!HGK?E@F!IJ!=G!DGIGHKE@GDZ!-GHGE@!RG!G[MUJHG! IBG!E@EIE?U!GMETG@GIEP!PB?@TGF!OE?!?PCIG!UJR:DJFG!?HFG@EIG!G[MJFCHGF!JQ!KJCFG! GK=H>J@EP!QE=HJ=U?FI!]<%2^!PGUUF!?@D!BEFIJ@G!-368_!KGIB>UIH?@FQGH?FG!;+(W1! N@JPNJCI!<%2!];JIW1:`:^!PGUUFZ! 2EHFIV!RG!DGPEDGD!IBG!G[MJFCHG!PJ@PG@IH?IEJ@!?@D!IEKG!JQ!FJDECK!?HFG@EIG! ?PPJHDE@T!IJ!IBG!G[MHGFFEJ@!JQ!&HQ7Z!9G!FI?HIGD!JCH!G[MJFCHG!G[MGHEKG@IF!REIB!IRJ! IEKG!MJE@IF!]a!?@D!7b!B^!?@D!QJCH!PJ@PG@IH?IEJ@F!]4V!7V!cV!?@D!W4!d<^Z!"QIGH!a!B! IHG?IKG@IV!IBG!?KJC@I!JQ!&HQ7!FBJRGD!DJFG:GQQGPI!HGU?IEJ@FBEMF!REIB!IBG! PJ@PG@IH?IEJ@F!JQ!FJDECK!?HFG@EIG!E@!=JIB!PGUU!UE@GF!?@D!IBGHG!RGHG!FET@EQEP?@I! DEQQGHG@PG!=GIRGG@!7!d<!?@D!c!d<!THJCMFZ!(BG!?KJC@I!JQ!&HQ7!?QIGH!7b!B!G[MJFCHG! ?UFJ!FBJRGD!DJFG:GQQGPI!HGU?IEJ@FBEMF!REIB!IBG!PJ@PG@IH?IEJ@F!JQ!FJDECK!?HFG@EIGZ! -JRGOGHV!PJKM?HGD!REIB!a!B!THJCMFV!7b!B!THJCMF!FBJRGD!DEQQGHG@I!M?IIGH@!JQ!&HQ7! ii G[MHGFFEJ@!=GIRGG@!IRJ!PGUU!UE@GFV!RBEPB!KETBI!=G!P?CFGD!=>!;JIW1!U?PNE@TZ! -

Histone Methylation by PRC2 Is Inhibited by Active Chromatin Marks
Histone methylation by PRC2 is inhibited by active chromatin marks Inauguraldissertation zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch–Naturwissenschaftlichen Fakultät der Universität Basel von Frank W. Schmitges aus Deutschland Basel, 2013 Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät auf Antrag von Prof. Dr. Susan Gasser, Dr. Nicolas Thomä, Dr. Wolfgang Fischle. Basel, den 20. September 2011 Prof. Dr. Susan Gasser (Fakultätsverantwortliche) Prof. Dr. Martin Spiess (Dekan) Table of Contents Abbreviations............................................................................................................................ 6 1. Summary ............................................................................................................................... 8 2. Introduction .......................................................................................................................... 9 2.1 Chromatin ......................................................................................................................... 9 2.2 Epigenetics ........................................................................................................................ 9 2.3 Mechanisms of epigenetic regulation ............................................................................. 10 2.3.1 DNA methylation ..................................................................................................... 11 2.3.2 Histone methylation ................................................................................................ -
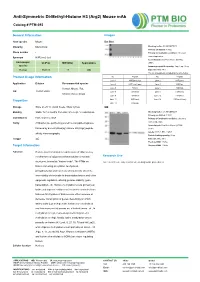
Anti-Symmetric Di-Methyl-Histone H3 (Arg2) Mouse Mab
Anti-Symmetric Di-Methyl-Histone H3 (Arg2) Mouse mAb Catalog # PTM-693 General Information Images Host species Mouse Dot Blot Clonality Monoclonal Blocking buffer: 5% NFDM/TBST Primary ab dilution: 1:1000 Clone number / Primary ab incubation condition: 2 hours at room temperature Synonym H3R2me2 (sy) Secondary ab: Goat Anti-Mouse IgG H&L Immunogen UniProt MW (kDa) Applications (HRP) species Immunogen peptide quantity: 1 ng, 4 ng, 16 ng Human P68431 17 WB Exposure time: 60 s The list of peptides is included in the table below: No. Peptide No. Peptide Product Usage Information Lane 1 H3R2me2 (sy) Lane 2 H3R2me1 Application Dilution Recommended species Lane 3 H3R2me2 (asy) Lane 4 H3K4ac Human, Mouse, Rat, Lane 5 H3K4cr Lane 6 H3K4hib WB 1:500-1:2000 Lane 7 H3K4me1 Lane 8 H3K4me2 Chlorocebus aethiops Lane 9 H3K4me3 Lane 10 H3R8me1 Lane 11 H3R8me2 Lane 12 H3R8me2 (asy) Properties Lane 13 H3R2un Storage Store at -20 °C. Avoid freeze / thaw cycles. WB Blocking buffer: 5% NFDM/TBST Stability Stable for 12 months from date of receipt / reconstitution Primary ab dilution: 1:2000 Constituents PBS, Glycerol, BSA Primary ab incubation condition: 2 hours at room temperature Purity Antibodies are purified by protein G-conjugated agarose Secondary ab: Goat Anti-Mouse IgG H&L followed by di-methylated(sy) histone H3 (Arg2) peptide (HRP) Lysate: MCF-7, BRL, COS-7 affinity chromatography. Protein loading quantity: 20 μg Isotype IgG Exposure time: 60 s Predicted MW: 17 kDa Target Information Observed MW: 17 kDa Function Histone post-translational modifications (PTMs) are key mechanisms of epigenetics that modulate chromatin Research Use structures, termed as “histone code”. -

Stepwise Histone Modifications Are Mediated by Multiple Enzymes That
ARTICLE Received 4 Apr 2013 | Accepted 29 Oct 2013 | Published 26 Nov 2013 DOI: 10.1038/ncomms3841 Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication Svetlana Petruk1, Kathryn L. Black1, Sina K. Kovermann2, Hugh W. Brock2 & Alexander Mazo1 The mechanism of epigenetic inheritance following DNA replication may involve dissociation of chromosomal proteins from parental DNA and reassembly on daughter strands in a specific order. Here we investigated the behaviour of different types of chromosomal proteins using newly developed methods that allow assessment of the assembly of proteins during DNA replication. Unexpectedly, most chromatin-modifying proteins tested, including methylases, demethylases, acetyltransferases and a deacetylase, are found in close proximity to PCNA or associate with short nascent DNA. Histone modifications occur in a temporal order following DNA replication, mediated by complex activities of different enzymes. In contrast, components of several major nucleosome-remodelling complexes are dissociated from parental DNA, and are later recruited to nascent DNA following replication. Epigenetic inheritance of gene expression patterns may require many aspects of chromatin structure to remain in close proximity to the replication complex followed by reassembly on nascent DNA shortly after replication. 1 Department of Biochemistry and Molecular Biology and Kimmel Cancer Center, Thomas Jefferson University, 1020 Locust Street, Philadelphia, Pennsylvania 19107, USA. 2 Department of Zoology, University of British Columbia, 6270 University Boulevard, Vancouver, British Columbia, Canada V6T 1Z4. Correspondence and requests for materials should be addressed to A.M. (email: [email protected]). NATURE COMMUNICATIONS | 4:2841 | DOI: 10.1038/ncomms3841 | www.nature.com/naturecommunications 1 & 2013 Macmillan Publishers Limited. -

H3r17me2(Asym)K18ac Polyclonal Antibody
H3R17me2(asym)K18ac polyclonal antibody Cat. No. C15410171 Specificity: Human, mouse, wide range expected Type: Polyclonal Purity: Affinity purified polyclonal antibody in PBS containing ChIP-grade / ChIP-seq-grade 0.05% azide and 0.05% ProClin 300. Source: Rabbit Storage: Store at -20°C; for long storage, store at -80°C. Lot #: A1922-001P Avoid multiple freeze-thaw cycles. Size: 50 µg/ 53 µl Precautions: This product is for research use only. Not for use in diagnostic or therapeutic procedures. Concentration: 0.94 µg/µl Description: Polyclonal antibody raised in rabbit against the region of histone H3 containing the asymmetrically dimethylated R17 and the acetylated lysine 18 (H3R17me2(asym)K18ac), using a KLH-conjugated synthetic peptide. Applications Suggested dilution Results ChIP* 1 µg per IP Fig 1, 2 ELISA 1:1,000 Fig 3 Dot blotting 1:20,000 Fig 4 Western blotting 1:1,000 Fig 5 IF 1:500 Fig 6 * Please note that the optimal antibody amount per ChIP should be determined by the end-user. We recommend testing 1-5 µg per IP. Target description Histones are the main constituents of the protein part of chromosomes of eukaryotic cells. They are rich in the amino acids arginine and lysine and have been greatly conserved during evolution. Histones pack the DNA into tight masses of chromatin. Two core histones of each class H2A, H2B, H3 and H4 assemble and are wrapped by 146 base pairs of DNA to form one octameric nucleosome. Histone tails undergo numerous post-translational modifications, which either directly or indirectly alter chromatin structure to facilitate transcriptional activation or repression or other nuclear processes. -

The Role of the Arginine Methyltransferase CARM1 in Global Transcriptional Regulation
Western University Scholarship@Western Electronic Thesis and Dissertation Repository 4-23-2014 12:00 AM The role of the arginine methyltransferase CARM1 in global transcriptional regulation. Niamh Coughlan The University of Western Ontario Supervisor Dr. Joseph Torchia The University of Western Ontario Graduate Program in Biochemistry A thesis submitted in partial fulfillment of the equirr ements for the degree in Doctor of Philosophy © Niamh Coughlan 2014 Follow this and additional works at: https://ir.lib.uwo.ca/etd Part of the Biochemistry Commons Recommended Citation Coughlan, Niamh, "The role of the arginine methyltransferase CARM1 in global transcriptional regulation." (2014). Electronic Thesis and Dissertation Repository. 2013. https://ir.lib.uwo.ca/etd/2013 This Dissertation/Thesis is brought to you for free and open access by Scholarship@Western. It has been accepted for inclusion in Electronic Thesis and Dissertation Repository by an authorized administrator of Scholarship@Western. For more information, please contact [email protected]. THE ROLE OF THE ARGININE METHYLTRANSFERASE CARM1 IN GLOBAL TRANSCRIPTIONAL REGULATION (Thesis format: Integrated Article) by Niamh Coughlan Graduate Program in Biochemistry A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy The School of Graduate and Postdoctoral Studies The University of Western Ontario London, Ontario, Canada © Niamh Coughlan 2014 Abstract Arginine methylation is a prevalent post-translational modification that is found on many nuclear and cytoplasmic proteins, and has been implicated in the regulation of gene expression. CARM1 is a member of the protein arginine methyltransferase (PRMT) family of proteins, and is a key protein responsible for arginine methylation of a subset of proteins involved in transcription.