Aneuraceae, Marchantiophytina)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Aquatic and Wet Marchantiophyta, Order Metzgeriales: Aneuraceae
Glime, J. M. 2021. Aquatic and Wet Marchantiophyta, Order Metzgeriales: Aneuraceae. Chapt. 1-11. In: Glime, J. M. Bryophyte 1-11-1 Ecology. Volume 4. Habitat and Role. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 11 April 2021 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 1-11: AQUATIC AND WET MARCHANTIOPHYTA, ORDER METZGERIALES: ANEURACEAE TABLE OF CONTENTS SUBCLASS METZGERIIDAE ........................................................................................................................................... 1-11-2 Order Metzgeriales............................................................................................................................................................... 1-11-2 Aneuraceae ................................................................................................................................................................... 1-11-2 Aneura .......................................................................................................................................................................... 1-11-2 Aneura maxima ............................................................................................................................................................ 1-11-2 Aneura mirabilis .......................................................................................................................................................... 1-11-7 Aneura pinguis .......................................................................................................................................................... -

Functional Gene Losses Occur with Minimal Size Reduction in the Plastid Genome of the Parasitic Liverwort Aneura Mirabilis
Functional Gene Losses Occur with Minimal Size Reduction in the Plastid Genome of the Parasitic Liverwort Aneura mirabilis Norman J. Wickett,* Yan Zhang, S. Kellon Hansen,à Jessie M. Roper,à Jennifer V. Kuehl,§ Sheila A. Plock, Paul G. Wolf,k Claude W. dePamphilis, Jeffrey L. Boore,§ and Bernard Goffinetà *Department of Ecology and Evolutionary Biology, University of Connecticut; Department of Biology, Penn State University; àGenome Project Solutions, Hercules, California; §Department of Energy Joint Genome Institute and University of California Lawrence Berkeley National Laboratory, Walnut Creek, California; and kDepartment of Biology, Utah State University Aneura mirabilis is a parasitic liverwort that exploits an existing mycorrhizal association between a basidiomycete and a host tree. This unusual liverwort is the only known parasitic seedless land plant with a completely nonphotosynthetic life history. The complete plastid genome of A. mirabilis was sequenced to examine the effect of its nonphotosynthetic life history on plastid genome content. Using a partial genomic fosmid library approach, the genome was sequenced and shown to be 108,007 bp with a structure typical of green plant plastids. Comparisons were made with the plastid genome of Marchantia polymorpha, the only other liverwort plastid sequence available. All ndh genes are either absent or pseudogenes. Five of 15 psb genes are pseudogenes, as are 2 of 6 psa genes and 2 of 6 pet genes. Pseudogenes of cysA, cysT, ccsA, and ycf3 were also detected. The remaining complement of genes present in M. polymorpha is present in the plastid of A. mirabilis with intact open reading frames. All pseudogenes and gene losses co-occur with losses detected in the plastid of the parasitic angiosperm Epifagus virginiana, though the latter has functional gene losses not found in A. -

A Taxonomic Revision of Aneuraceae (Marchantiophyta) from Eastern Africa with an Interactive Identification Key
cryptogamie Bryologie 2019 ● 41 ● 2 DIRECTEUR DE LA PUBLICATION : Bruno David, Président du Muséum national d’Histoire naturelle RÉDACTEURS EN CHEF / EDITORS-IN-CHIEF : Denis LAMY, Michelle Price ASSISTANTS DE RÉDACTION / ASSISTANT EDITORS : Marianne SALAÜN ([email protected]) MISE EN PAGE / PAGE LAYOUT : Marianne SALAÜN RÉDACTEURS ASSOCIÉS / ASSOCIATE EDITORS Biologie moléculaire et phylogénie / Molecular biology and phylogeny Bernard GOFFINET Department of Ecology and Evolutionary Biology, University of Connecticut (United States) Mousses d’Europe / European mosses Isabel DRAPER Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) Francisco LARA GARCÍA Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) Mousses d’Afrique et d’Antarctique / African and Antarctic mosses Rysiek OCHYRA Laboratory of Bryology, Institute of Botany, Polish Academy of Sciences, Krakow (Pologne) Bryophytes d’Asie / Asian bryophytes Rui-Liang ZHU School of Life Science, East China Normal University, Shanghai (China) Bioindication / Biomonitoring Franck-Olivier DENAYER Faculté des Sciences Pharmaceutiques et Biologiques de Lille, Laboratoire de Botanique et de Cryptogamie, Lille (France) Écologie des bryophytes / Ecology of bryophyte Nagore GARCÍA MEDINA Department of Biology (Botany), and Centro de Investigación en Biodiversidad y Cambio Global (CIBC-UAM), Universidad Autónoma de Madrid (Spain) COUVERTURE / COVER : From top left, to bottom right, by -

Article ISSN 1179-3163 (Online Edition)
Phytotaxa 63: 21–68 (2012) ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ PHYTOTAXA Copyright © 2012 Magnolia Press Article ISSN 1179-3163 (online edition) Early Land Plants Today: Index of Liverworts & Hornworts 2009–2010 LARS SÖDERSTRÖM1, ANDERS HAGBORG2, MARSHALL R. CROSBY3 & MATT VON KONRAT2 1 Department of Biology, Norwegian University of Science and Technology, N-7491, Trondheim, Norway; [email protected] 2 Department of Botany, The Field Museum, 1400 South Lake Shore Drive, Chicago, IL 60605–2496, U.S.A.;[email protected], [email protected] 3 Missouri Botanical Garden, P. O. Box 299, St. Louis, MO 63166–0299 U.S.A.; [email protected] Abstract A widely accessible list of known plant species is a fundamental requirement for plant conservation and has vast applications. An index of published names of liverworts and hornworts between 2009 and 2010 is provided as part of a continued effort in working toward producing a world checklist of this group. Included in the list are also names overlooked by earlier indices. The list includes 30 higher taxa, 250 species, 52 infraspecific taxa, 31 autonyms, and two fossils for 2009 and 2010. A number of taxa not covered by the earlier indices for 2000-2008 are also included. Key words: Liverworts, hornworts, index, nomenclature Introduction Under the auspices of the Early Land Plants Today project, there has been a strong community-driven effort attempting to address the critical need to synthesize the vast nomenclatural, taxonomic and global distributional data for liverworts (Marchantiophyta) and hornworts (Anthocerotophyta) (von Konrat et al. 2010a). These endeavours are critical in providing the foundation to develop a working checklist of liverworts and hornworts worldwide; the first version is projected to be published in 2012. -

Bryophyte Ecology Table of Contents
Glime, J. M. 2020. Table of Contents. Bryophyte Ecology. Ebook sponsored by Michigan Technological University 1 and the International Association of Bryologists. Last updated 15 July 2020 and available at <https://digitalcommons.mtu.edu/bryophyte-ecology/>. This file will contain all the volumes, chapters, and headings within chapters to help you find what you want in the book. Once you enter a chapter, there will be a table of contents with clickable page numbers. To search the list, check the upper screen of your pdf reader for a search window or magnifying glass. If there is none, try Ctrl G to open one. TABLE OF CONTENTS BRYOPHYTE ECOLOGY VOLUME 1: PHYSIOLOGICAL ECOLOGY Chapter in Volume 1 1 INTRODUCTION Thinking on a New Scale Adaptations to Land Minimum Size Do Bryophytes Lack Diversity? The "Moss" What's in a Name? Phyla/Divisions Role of Bryology 2 LIFE CYCLES AND MORPHOLOGY 2-1: Meet the Bryophytes Definition of Bryophyte Nomenclature What Makes Bryophytes Unique Who are the Relatives? Two Branches Limitations of Scale Limited by Scale – and No Lignin Limited by Scale – Forced to Be Simple Limited by Scale – Needing to Swim Limited by Scale – and Housing an Embryo Higher Classifications and New Meanings New Meanings for the Term Bryophyte Differences within Bryobiotina 2-2: Life Cycles: Surviving Change The General Bryobiotina Life Cycle Dominant Generation The Life Cycle Life Cycle Controls Generation Time Importance Longevity and Totipotency 2-3: Marchantiophyta Distinguishing Marchantiophyta Elaters Leafy or Thallose? Class -

Wikstrom2009chap13.Pdf
Liverworts (Marchantiophyta) Niklas Wikströma,*, Xiaolan He-Nygrénb, and our understanding of phylogenetic relationships among A. Jonathan Shawc major lineages and the origin and divergence times of aDepartment of Systematic Botany, Evolutionary Biology Centre, those lineages. Norbyvägen 18D, Uppsala University, Norbyvägen 18D 75236, Altogether, liverworts (Phylum Marchantiophyta) b Uppsala, Sweden; Botanical Museum, Finnish Museum of Natural comprise an estimated 5000–8000 living species (8, 9). History, University of Helsinki, P.O. Box 7, 00014 Helsinki, Finland; Early and alternative classiA cations for these taxa have cDepartment of Biology, Duke University, Durham, NC 27708, USA *To whom correspondence should be addressed (niklas.wikstrom@ been numerous [reviewed by Schuster ( 10)], but the ebc.uu.se) arrangement of terminal taxa (species, genera) into lar- ger groups (e.g., families and orders) based on morpho- logical criteria alone began in the 1960s and 1970s with Abstract the work of Schuster (8, 10, 11) and Schljakov (12, 13), and culminated by the turn of the millenium with the work Liverworts (Phylum Marchantiophyta) include 5000–8000 of Crandall-Stotler and Stotler (14). 7 ree morphological species. Phylogenetic analyses divide liverworts into types of plant bodies (gametophytes) have generally been Haplomitriopsida, Marchantiopsida, and Jungerman- recognized and used in liverwort classiA cations: “com- niopsida. Complex thalloids are grouped with Blasiales in plex thalloids” including ~6% of extant species diversity Marchantiopsida, and leafy liverworts are grouped with and with a thalloid gametophyte that is organized into Metzgeriidae and Pelliidae in Jungermanniopsida. The distinct layers; “leafy liverworts”, by far the most speci- timetree shows an early Devonian (408 million years ago, ose group, including ~86% of extant species diversity and Ma) origin for extant liverworts. -

Hepaticae, Metzgeriales)
Cryptogamie, Bryologie, 2010, 31 (3): 207-215 ©2010 Adac. Tous droits réservés Fast identification method for an allopolyploid liverwort Pellia borealis Lorbeer (Hepaticae, Metzgeriales) Ewa CHUDZI◊SKA*&Ireneusz J. ODRZYKOSKI Department of Genetics, Institute of Experimental Biology, Adam Mickiewicz University, Umultowska 89, 61-614 Pozna¬,Poland (Received 13 February 2009, accepted 22 March 2010) Abstract –Asimple method was described for the identification of an allopolyploid liverwort Pellia borealis based on isozyme analysis. Aperfect correlation was observed between the polyploid chromosome number (n=18) and the fixed heterozygous isoenzyme pattern of two diaphorase loci (DiaA and DiaC) in alarge collection of Pellia epiphylla sensu lato from Polish populations. Therefore, their isozyme pattern may be used as a reliable diagnostic marker, which makes it possible to distinguish between apolyploid and two parental cryptic species from the P. epiphylla complex. Due to the alloploid origin of P. borealis we postulate that the species rank of the taxon should be maintained and geographical distribution revised. Isozyme markers /chromosome number /allopolyploidy / Pellia borealis /Pellia epiphylla complex INTRODUCTION Liverworts from genus Pellia the Epiphyllae section (Schust.) occurs in North America, Europe and Asia (de Sloover &Messe, 1982; Newton, 1986a; Szweykowski et al., 1995; Odrzykoski et al., 1996). Recent studies on genetic variability of this section have provided new evidence supporting ahypothesis of allopolyploid origins of Pellia borealis Lorbeer as aspecies originated by hybridization between two cryptic species from the Pellia epiphylla (L.) Corda complex and subsequent polyploidization (Odrzykoski et al.,1996; Chudzi¬ska et al., 2005; Fiedorow et al.,2001; Pacak &Szweykowska-Kuli¬ska, 2000, 2003). -
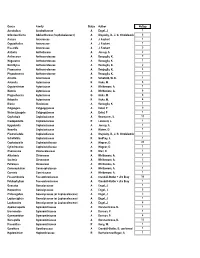
Family Status Author Nospp Acrobolbus Acrobolbaceae a Engel, J
Genus Family Status Author NoSpp Acrobolbus Acrobolbaceae A Engel, J. 1 Odontoschisma Adelanthaceae (Cephaloziaceae?) A Krayesky, D., J. G. Chmielewski 6 Aneura Aneuraceae A J. Faubert 1 Cryptothallus Aneuraceae A J. Faubert 1 Riccardia Aneuraceae A J. Faubert 5 Anthelia Antheliaceae A Jessup, S. 2 Anthoceros Anthocerotaceae A Renzaglia, K. 7 Megaceros Anthocerotaceae A Renzaglia, K. 1 Notothylas Anthocerotaceae A Renzaglia, K. 2 Phaeoceros Anthocerotaceae A Renzaglia, K. 4 Phymatoceros Anthocerotaceae A Renzaglia, K. 1 Arnellia Arnelliaceae R Schofield, W. B. 1 Asterella Aytoniaceae R Hicks, M. 8 Cryptomitrium Aytoniaceae A Whittemore, A. 1 Mannia Aytoniaceae A Whittemore, A. 6 Plagiochasma Aytoniaceae G Hicks, M. 6 Reboulia Aytoniaceae R Hicks, M. 4 Blasia Blasiaceae A Renzaglia, K. 1 Calypogeia Calypogejaceae A Eckel, P 9 Metacalypogeia Calypogejaceae A Eckel, P 1 Cephalozia Cephaloziaceae A Newmaster, S. 11 Cladopodiella Cephaloziaceae R Leonardi, L. 1 Hygrobiella Cephaloziaceae A Jessup, S. 1 Nowellia Cephaloziaceae A Waters, D. 1 Pleurocladula Cephaloziaceae A Krayesky, D., J. G. Chmielewski 1 Schofieldia Cephaloziaceae R Godfrey, J. 1 Cephaloziella Cephaloziellaceae A Wagner, D. 21 Cylindrocolea Cephaloziellaceae A Wagner, D. 1 Chonecolea Chonecoleaceae R Breil, D. 1 Athalamia Cleveaceae A Whittemore, A. 1 Sauteria Cleveaceae A Whittemore, A. 2 Peltolepis Cleveaceae A Whittemore, A. 1 Conocephalum Conocephalaceae A Whittemore, A. 1 Corsinia Corsiniaceae A Whittemore, A. 1 Fossombronia Fossombroniaceae A Crandall-Stotler + Jim Bray 10 Petalophyllum Fossombroniaceae A Crandall-Stotler + Jim Bray 1 Geocalyx Geocalycaceae A Engel, J. 1 Harpanthus Geocalycaceae A Engel, J. 3 Chiloscyphus Geocalycaceae (or Lophocoleaceae) A Engel, J. 3 Leptoscyphus Geocalycaceae (or Lophocoleaceae) A Engel, J. -

BRYOPHYTES .Pdf
Diversity of Microbes and Cryptogams Bryophyta Geeta Asthana Department of Botany, University of Lucknow, Lucknow – 226007 India Date of submission: May 11, 2006 Version: English Significant Key words: Bryophyta, Hepaticopsida (Liverworts), Anthocerotopsida (Hornworts), , Bryopsida (Mosses). 1 Contents 1. Introduction • Definition & Systematic Position in the Plant Kingdom • Alternation of Generation • Life-cycle Pattern • Affinities with Algae and Pteridophytes • General Characters 2. Classification 3. Class – Hepaticopsida • General characters • Classification o Order – Calobryales o Order – Jungermanniales – Frullania o Order – Metzgeriales – Pellia o Order – Monocleales o Order – Sphaerocarpales o Order – Marchantiales – Marchantia 4. Class – Anthocerotopsida • General Characters • Classification o Order – Anthocerotales – Anthoceros 5. Class – Bryopsida • General Characters • Classification o Order – Sphagnales – Sphagnum o Order – Andreaeales – Andreaea o Order – Takakiales – Takakia o Order – Polytrichales – Pogonatum, Polytrichum o Order – Buxbaumiales – Buxbaumia o Order – Bryales – Funaria 6. References 2 Introduction Bryophytes are “Avascular Archegoniate Cryptogams” which constitute a large group of highly diversified plants. Systematic position in the plant kingdom The plant kingdom has been classified variously from time to time. The early systems of classification were mostly artificial in which the plants were grouped for the sake of convenience based on (observable) evident characters. Carolus Linnaeus (1753) classified -

Downloads/figure(Reversal Cost).Svg 1/1
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.08.430207; this version posted February 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 Living apart if you can – how genetically and developmentally controlled sex has shaped the 2 evolution of liverworts 3 4 Xiaolan He1, Jorge R. Flores1, Yu Sun2, John L. Bowman3 5 6 1.Finnish Museum of Natural History, University of Helsinki, FI-00014, Helsinki, Finland 7 2. Lushan Botanical Garden, Chinese Academy of Sciences, Jiujiang, Jiangxi 332900, China 8 3. School of Biological Science, Monash University, Melbourne VIC 3800, Australia 9 10 11 12 Abstract 13 Sexual differentiation in bryophytes occurs in the dominant gametophytic generation. Over half of 14 bryophytes are dioicous, and this pattern in liverworts is even more profound as over 70% of 15 species are dioicous. However, the evolutionary mechanisms leading to the prevalence of dioicy 16 and the shifts of sexual systems between dioicy and monoicy have remained poorly known. These 17 essential factors in reproductive biology are explored here in light of phylogenetics combined with 18 evidence of genomic characterization of sex chromosomes and sex-determination, as well as 19 cytology. Our analyses and discussions on liverworts are focused on: (1) ancestry and shifts in 20 sexuality, (2) evolution of sex chromosomes and maintenance of haploid dioicy, and (3) 21 environmental impact on the evolution of monoicism. -

Sites of Importance for Nature Conservation Wales Guidance (Pdf)
Wildlife Sites Guidance Wales A Guide to Develop Local Wildlife Systems in Wales Wildlife Sites Guidance Wales A Guide to Develop Local Wildlife Systems in Wales Foreword The Welsh Assembly Government’s Environment Strategy for Wales, published in May 2006, pays tribute to the intrinsic value of biodiversity – ‘the variety of life on earth’. The Strategy acknowledges the role biodiversity plays, not only in many natural processes, but also in the direct and indirect economic, social, aesthetic, cultural and spiritual benefits that we derive from it. The Strategy also acknowledges that pressures brought about by our own actions and by other factors, such as climate change, have resulted in damage to the biodiversity of Wales and calls for a halt to this loss and for the implementation of measures to bring about a recovery. Local Wildlife Sites provide essential support between and around our internationally and nationally designated nature sites and thus aid our efforts to build a more resilient network for nature in Wales. The Wildlife Sites Guidance derives from the shared knowledge and experience of people and organisations throughout Wales and beyond and provides a common point of reference for the most effective selection of Local Wildlife Sites. I am grateful to the Wales Biodiversity Partnership for developing the Wildlife Sites Guidance. The contribution and co-operation of organisations and individuals across Wales are vital to achieving our biodiversity targets. I hope that you will find the Wildlife Sites Guidance a useful tool in the battle against biodiversity loss and that you will ensure that it is used to its full potential in order to derive maximum benefit for the vitally important and valuable nature in Wales. -

New Insights Into the Phylogeny and Relationships Within the Worldwide Genus Riccardia (Aneuraceae, Marchantiophytina)
European Journal of Taxonomy 273: 1–26 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2017.273 www.europeanjournaloftaxonomy.eu 2017 · Rabeau L. et al. This work is licensed under a Creative Commons Attribution 3.0 License. DNA Library of Life, research article New insights into the phylogeny and relationships within the worldwide genus Riccardia (Aneuraceae, Marchantiophytina) Lucile RABEAU 1,*, S. Robbert GRADSTEIN 2, Jean-Yves DUBUISSON 3, Martin NEBEL 4, Dietmar QUANDT 5 & Catherine REEB 6 1,2,3,6 Université Pierre et Marie Curie – Sorbonne Universités – Institut de Systématique, Évolution, Biodiversité, ISYEB, UMR 7205, CNRS-MNHN-UPMC-EPHE, Muséum national d’Histoire naturelle, 57 rue Cuvier, CP 39, 75005 Paris, France. 4 Staatliches Museum für Naturkunde Stuttgart, Rosenstein 1, 70191 Stuttgart, Germany. 5 Nees-Institut für Biodiversität der Pflanzen, Rheinische Friedrich-Wilhelms-Universität Bonn, Meckenheimer Allee 170, 53115 Bonn, Germany. * Corresponding author: [email protected] 2 Email: [email protected] 3 Email: [email protected] 4 Email: [email protected] 5 Email: [email protected] 6 Email: [email protected] Abstract. With 280 accepted species, the genus Riccardia S.F.Gray (Aneuraceae) is one of the most speciose genera of simple thalloid liverworts. The current classification of this genus is based on morphological and limited-sampling molecular studies. Very few molecular data are available and a comprehensive view of evolutionary relationships within the genus is still lacking. A phylogeny focusing on relationships within the large genus Riccardia has not been conducted. Here we propose the first worldwide molecular phylogeny of the genus Riccardia, based on Bayesian inference and parsimony ratchet analyses of sequences from three plastid regions (psbA-trnH, rps4, trnL-F).