S41467-018-03432-4.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Imm Catalog.Pdf
$ Gene Symbol A B 3 C 4 D 9 E 10 F 11 G 12 H 13 I 14 J. K 17 L 18 M 19 N 20 O. P 22 R 26 S 27 T 30 U 32 V. W. X. Y. Z 33 A ® ® Gene Symbol Gene ID Antibody Monoclonal Antibody Polyclonal MaxPab Full-length Protein Partial-length Protein Antibody Pair KIt siRNA/Chimera Gene Symbol Gene ID Antibody Monoclonal Antibody Polyclonal MaxPab Full-length Protein Partial-length Protein Antibody Pair KIt siRNA/Chimera A1CF 29974 ● ● ADAMTS13 11093 ● ● ● ● ● A2M 2 ● ● ● ● ● ● ADAMTS20 80070 ● AACS 65985 ● ● ● ADAMTS5 11096 ● ● ● AANAT 15 ● ● ADAMTS8 11095 ● ● ● ● AATF 26574 ● ● ● ● ● ADAMTSL2 9719 ● AATK 9625 ● ● ● ● ADAMTSL4 54507 ● ● ABCA1 19 ● ● ● ● ● ADAR 103 ● ● ABCA5 23461 ● ● ADARB1 104 ● ● ● ● ABCA7 10347 ● ADARB2 105 ● ABCB9 23457 ● ● ● ● ● ADAT1 23536 ● ● ABCC4 10257 ● ● ● ● ADAT2 134637 ● ● ABCC5 10057 ● ● ● ● ● ADAT3 113179 ● ● ● ABCC8 6833 ● ● ● ● ADCY10 55811 ● ● ABCD2 225 ● ADD1 118 ● ● ● ● ● ● ABCD4 5826 ● ● ● ADD3 120 ● ● ● ABCG1 9619 ● ● ● ● ● ADH5 128 ● ● ● ● ● ● ABL1 25 ● ● ADIPOQ 9370 ● ● ● ● ● ABL2 27 ● ● ● ● ● ADK 132 ● ● ● ● ● ABO 28 ● ● ADM 133 ● ● ● ABP1 26 ● ● ● ● ● ADNP 23394 ● ● ● ● ABR 29 ● ● ● ● ● ADORA1 134 ● ● ACAA2 10449 ● ● ● ● ADORA2A 135 ● ● ● ● ● ● ● ACAN 176 ● ● ● ● ● ● ADORA2B 136 ● ● ACE 1636 ● ● ● ● ADRA1A 148 ● ● ● ● ACE2 59272 ● ● ADRA1B 147 ● ● ACER2 340485 ● ADRA2A 150 ● ● ACHE 43 ● ● ● ● ● ● ADRB1 153 ● ● ACIN1 22985 ● ● ● ADRB2 154 ● ● ● ● ● ACOX1 51 ● ● ● ● ● ADRB3 155 ● ● ● ● ACP5 54 ● ● ● ● ● ● ● ADRBK1 156 ● ● ● ● ACSF2 80221 ● ● ADRM1 11047 ● ● ● ● ACSF3 197322 ● ● AEBP1 165 ● ● ● ● ACSL4 2182 ● -

Datasheet Blank Template
SAN TA C RUZ BI OTEC HNOL OG Y, INC . LRP3 (E-13): sc-109956 BACKGROUND APPLICATIONS Members of the LDL receptor gene family, including LDLR (low density lipo- LRP3 (E-13) is recommended for detection of All LRP3 isoforms 1-3 of mouse, protein receptor), LRP1 (low density lipoprotein related protein), Megalin rat and human origin by Western Blotting (starting dilution 1:200, dilution (also designated GP330), VLDLR (very low density lipoprotein receptor) and range 1:100-1:1000), immunofluorescence (starting dilution 1:50, dilution ApoER2 are characterized by a cluster of cysteine-rich class A repeats, epi - range 1:50-1:500) and solid phase ELISA (starting dilution 1:30, dilution dermal growth factor (EGF)-like repeats, YWTD repeats and an O-linked sugar range 1:30-1:3000); non cross-reactive with other LRP family members. domain. Low-density lipoprotein receptor-related protein 3 (LRP3) is a 770 LRP3 (E-13) is also recommended for detection of All LRP3 isoforms 1-3 in amino acid protein that contains two CUB domains and four LDL-receptor additional species, including equine, canine, bovine and porcine. class A domains. LRP3 is widely expressed with highest expression in skele - tal muscle and ovary and lowest expression in testis, colon and leukocytes. Suitable for use as control antibody for LRP3 siRNA (h): sc-97441, LRP3 LRP3 is potentially a membrane receptor involved in the internalization of siRNA (m): sc-149048, LRP3 shRNA Plasmid (h): sc-97441-SH, LRP3 shRNA lipophilic molecules and/or signal transduction. Plasmid (m): sc-149048-SH, LRP3 shRNA (h) Lentiviral Particles: sc-97441-V and LRP3 shRNA (m) Lentiviral Particles: sc-149048-V. -

Prevention & Control of Viral Hepatitis Infection
Prevention & Control of Viral Hepatitis Infection: A Strategy for Global Action © World Health Organization 2011. All rights reserved. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. All reasonable precautions have been taken by WHO to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either express or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall the World Health Organization be liable for damages arising from its use. Table of contents Disease burden 02 What is viral hepatitis? 05 Prevention & control: a tailored approach 06 Global Achievements 08 Remaining challenges 10 World Health Assembly: a mandate for comprehensive prevention & control 13 WHO goals and strategy -

The Role of Hepatitis C Virus in Hepatocellular Carcinoma U
Viruses in cancer cell plasticity: the role of hepatitis C virus in hepatocellular carcinoma U. Hibner, D. Gregoire To cite this version: U. Hibner, D. Gregoire. Viruses in cancer cell plasticity: the role of hepatitis C virus in hepato- cellular carcinoma. Contemporary Oncology, Termedia Publishing House, 2015, 19 (1A), pp.A62–7. 10.5114/wo.2014.47132. hal-02187396 HAL Id: hal-02187396 https://hal.archives-ouvertes.fr/hal-02187396 Submitted on 2 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - ShareAlike| 4.0 International License Review Viruses are considered as causative agents of a significant proportion of human cancers. While the very Viruses in cancer cell plasticity: stringent criteria used for their clas- sification probably lead to an under- estimation, only six human viruses the role of hepatitis C virus are currently classified as oncogenic. In this review we give a brief histor- in hepatocellular carcinoma ical account of the discovery of on- cogenic viruses and then analyse the mechanisms underlying the infectious causes of cancer. We discuss viral strategies that evolved to ensure vi- Urszula Hibner1,2,3, Damien Grégoire1,2,3 rus propagation and spread can alter cellular homeostasis in a way that increases the probability of oncogen- 1Institut de Génétique Moléculaire de Montpellier, CNRS, UMR 5535, Montpellier, France ic transformation and acquisition of 2Université Montpellier 2, Montpellier, France stem cell phenotype. -

Rational Engineering of HCV Vaccines for Humoral Immunity
viruses Review To Include or Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity Felicia Schlotthauer 1,2,†, Joey McGregor 1,2,† and Heidi E Drummer 1,2,3,* 1 Viral Entry and Vaccines Group, Burnet Institute, Melbourne, VIC 3004, Australia; [email protected] (F.S.); [email protected] (J.M.) 2 Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC 3000, Australia 3 Department of Microbiology, Monash University, Clayton, VIC 3800, Australia * Correspondence: [email protected]; Tel.: +61-392-822-179 † These authors contributed equally to this work. Abstract: Direct-acting antiviral agents have proven highly effective at treating existing hepatitis C infections but despite their availability most countries will not reach the World Health Organization targets for elimination of HCV by 2030. A prophylactic vaccine remains a high priority. Whilst early vaccines focused largely on generating T cell immunity, attention is now aimed at vaccines that gen- erate humoral immunity, either alone or in combination with T cell-based vaccines. High-resolution structures of hepatitis C viral glycoproteins and their interaction with monoclonal antibodies isolated from both cleared and chronically infected people, together with advances in vaccine technologies, provide new avenues for vaccine development. Keywords: glycoprotein E2; vaccine development; humoral immunity Citation: Schlotthauer, F.; McGregor, J.; Drummer, H.E To Include or 1. Introduction Occlude: Rational Engineering of HCV Vaccines for Humoral Immunity. The development of direct-acting antiviral agents (DAA) with their ability to cure Viruses 2021, 13, 805. https:// infection in >95% of those treated was heralded as the key to eliminating hepatitis C globally. -
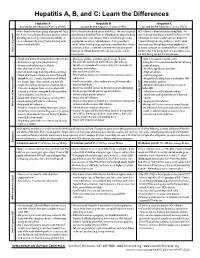
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

Understanding Human Astrovirus from Pathogenesis to Treatment
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 6-2020 Understanding Human Astrovirus from Pathogenesis to Treatment Virginia Hargest University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Diseases Commons, Medical Sciences Commons, and the Viruses Commons Recommended Citation Hargest, Virginia (0000-0003-3883-1232), "Understanding Human Astrovirus from Pathogenesis to Treatment" (2020). Theses and Dissertations (ETD). Paper 523. http://dx.doi.org/10.21007/ etd.cghs.2020.0507. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Understanding Human Astrovirus from Pathogenesis to Treatment Abstract While human astroviruses (HAstV) were discovered nearly 45 years ago, these small positive-sense RNA viruses remain critically understudied. These studies provide fundamental new research on astrovirus pathogenesis and disruption of the gut epithelium by induction of epithelial-mesenchymal transition (EMT) following astrovirus infection. Here we characterize HAstV-induced EMT as an upregulation of SNAI1 and VIM with a down regulation of CDH1 and OCLN, loss of cell-cell junctions most notably at 18 hours post-infection (hpi), and loss of cellular polarity by 24 hpi. While active transforming growth factor- (TGF-) increases during HAstV infection, inhibition of TGF- signaling does not hinder EMT induction. However, HAstV-induced EMT does require active viral replication. -

Host Cell Functions in Vesicular Stomatitis Virus Replication Phat X
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Veterinary and Biomedical Science Veterinary and Biomedical Sciences, Department of 2016 Host Cell Functions In Vesicular Stomatitis Virus Replication Phat X. Dinh University of Nebraska - Lincoln Anshuman Das University of Nebraska-Lincoln Asit K. Pattnaik University of Nebraska - Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/vetscipapers Part of the Biochemistry, Biophysics, and Structural Biology Commons, Cell and Developmental Biology Commons, Immunology and Infectious Disease Commons, Medical Sciences Commons, Veterinary Microbiology and Immunobiology Commons, and the Veterinary Pathology and Pathobiology Commons Dinh, Phat X.; Das, Anshuman; and Pattnaik, Asit K., "Host Cell Functions In Vesicular Stomatitis Virus Replication" (2016). Papers in Veterinary and Biomedical Science. 237. https://digitalcommons.unl.edu/vetscipapers/237 This Article is brought to you for free and open access by the Veterinary and Biomedical Sciences, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers in Veterinary and Biomedical Science by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published in Biology and Pathogenesis of Rhabdo and Filoviruses Copyright © 2016 World Scientific Publishing Co Pte Ltd digitalcommons.unl.edu HOST CELL FUNCTIONS IN VESICULAR STOMATITIS VIRUS REPLICATION Phat X. Dinh, Anshuman Das, and Asit K. Pattnaik School of Veterinary Medicine and Biomedical Sciences and the Nebraska Center for Virology, University of Nebraska-Lincoln, 109 Morrison Life Science Research Center, 4240 Fair Street, Lincoln, * Nebraska 68583, USA E-mail: [email protected] Summary Vesicular stomatitis virus (VSV), the prototypic rhabdovirus, has been used as an excellent paradigm for understanding the mechanisms of virus replication, pathogenesis, host response to virus infection and also for myriads of studies on cellular and molecular biology. -

The Role of Hepatitis C Virus in Hepatocellular Carcinoma A63
Review Viruses are considered as causative agents of a significant proportion of human cancers. While the very Viruses in cancer cell plasticity: stringent criteria used for their clas- sification probably lead to an under- estimation, only six human viruses the role of hepatitis C virus are currently classified as oncogenic. In this review we give a brief histor- in hepatocellular carcinoma ical account of the discovery of on- cogenic viruses and then analyse the mechanisms underlying the infectious causes of cancer. We discuss viral strategies that evolved to ensure vi- Urszula Hibner1,2,3, Damien Grégoire1,2,3 rus propagation and spread can alter cellular homeostasis in a way that increases the probability of oncogen- 1Institut de Génétique Moléculaire de Montpellier, CNRS, UMR 5535, Montpellier, France ic transformation and acquisition of 2Université Montpellier 2, Montpellier, France stem cell phenotype. We argue that 3Université Montpellier 1, Montpellier, France a useful way of analysing the conver- gent characteristics of viral infection and cancer is to examine how viruses affect the so-called cancer hallmarks. Introduction This view of infectious origin of cancer It is estimated that close to 20% of human cancers are due to infections is illustrated by examples from hep- atitis C infection, which is associated with known pathogens, mainly with viruses [1]. Given the difficulties in un- with a high proportion of hepatocellu- ambiguously assigning their causative role [2] and the fast rate of discovery lar carcinoma. of new viruses [3], it is likely a conservative estimate. Different pathogens are associated with increased cancer risk. One of the Key words: viruses, cancer cell plas- first identified cancer-causing infectious disease was schistosomiasis, ini- ticity, HCV, hepatocellular carcinoma. -

Revisiting the Elusive Hepatitis C Vaccine
Editorial Revisiting the Elusive Hepatitis C Vaccine Stephen M. Todryk 1,2,* , Margaret F. Bassendine 2,* and Simon H. Bridge 1,2,* 1 Faculty of Health & Life Sciences, Northumbria University, Newcastle upon Tyne NE1 8ST, UK 2 Translational & Clinical Research Institute, The Medical School, Newcastle University, Newcastle upon Tyne NE2 4HH, UK * Correspondence: [email protected] (S.M.T.); [email protected] (M.F.B.); [email protected] (S.H.B.) The impactful discovery and subsequent characterisation of hepatitis C virus (HCV), an RNA virus of the flavivirus family, led to the awarding of the 2020 Nobel Prize in Physiology or Medicine to Harvey J. Alter, Michael Houghton and Charles M. Rice [1]. However, despite the significant advances recognised by this Nobel prize, an effective HCV vaccine remains elusive. The recent success of vaccines against SARS-CoV-2, developed with unprecedented speed, has shone a bright light on the vaccination process for protection against viral threats and may provide renewed impetus for the development of vaccines for other viruses. HCV infection remains a major global health problem as approximately three quarters of those infected develop chronic infection, leading to morbidity and mortality due to progressive liver fibrosis, cirrhosis and cancer. The World Health Organization (WHO) estimates that 71.1 million people are living with chronic HCV, resulting in over 400,000 deaths every year. In 2016, the WHO adopted a global strategy with the aim of eliminating viral hepatitis as a public health problem, comprising targets to reduce new viral hepatitis infections by 90% and reduce deaths due to viral hepatitis by 65% by 2030. -

Megalin, an Endocytotic Receptor with Signalling Potential
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 116 Megalin, an Endocytotic Receptor with Signalling Potential MÅRTEN LARSSON ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6206 UPPSALA ISBN 91-554-6483-1 2006 urn:nbn:se:uu:diva-6585 !""# $% & ' & & ( ) & *+ , - . '+ / + !""#+ ' . 0 - 1' ' ( + 2 + #+ #" + + 314 56%%6#7 6+ ' ' ' -6 & + 3 & - ' & + 0 - 8 ' & ' ' + , & ' ' - 6 ' - - & ' + 2 ' && ' 5% )(165%* - & ' - (96 + 6 : ;.<6!5 8 & + , (165% (165 12("! - & ' - (9!6 ' & 12(5= + ' 2 , - & + ' - )41* - + 4 ' 8 - ' + , 6 & ' ' & - 8 + ' - & & + & '' ' ' & + 3 ' ' - 6 - + , '' ' & & ' ' - + ' ' - & ' ' - 8 68 ' ' - + 8 & ' )0(.* ' ' && + 1 ' & ' 0(. ' 8 - ;6 6 ' 8 ' - //6 & & ' 6 )02(* - ' & 8 & 0(.+ ' /0(6! ( 65% 0 6 ' >' ! " # $ " # % &'(" " )*+&,(- " ? @ / !""# 3114 #%6#!"# 314 56%%6#7 6 $ $$$ 6#%7% ) $AA +8+A B C $ $$$ 6#%7%* To Dr. John Pemberton List of original papers This thesis is based on the following -

Microrna-4739 Regulates Osteogenic and Adipocytic Differentiation of Immortalized Human Bone Marrow Stromal Cells Via Targeting LRP3
Stem Cell Research 20 (2017) 94–104 Contents lists available at ScienceDirect Stem Cell Research journal homepage: www.elsevier.com/locate/scr MicroRNA-4739 regulates osteogenic and adipocytic differentiation of immortalized human bone marrow stromal cells via targeting LRP3 Mona Elsafadi a,c, Muthurangan Manikandan a, Nehad M Alajez a,RimiHamama, Raed Abu Dawud b, Abdullah Aldahmash a,d,ZafarIqbale,MusaadAlfayeza,MoustaphaKassema,c,AmerMahmooda,⁎ a Stem Cell Unit, Department of Anatomy, College of Medicine,King Saud University, Riyadh 11461, Saudi Arabia b Department of Comparative Medicine, King Faisal Specialist Hospital and Research Centre, Riyadh 12713, Saudi Arabia c KMEB, Department of Endocrinology, University Hospital of Odense, University of Southern Denmark, Winslowsparken 25.1, DK-5000 Odense C, Denmark d Prince Naif Health Research Center, King Saud University, Riyadh 11461, Saudi Arabia e Department of Basic Sciences, College of applied medical sciences, King Saud Bin Abdulaziz University for Health Sciences (KSAU-HS), National GuardHealthAffairs,AlAhsa,SaudiArabia article info abstract Article history: Understanding the regulatory networks underlying lineage differentiation and fate determination of human Received 7 September 2016 bone marrow stromal cells (hBMSC) is a prerequisite for their therapeutic use. The goal of the current study Received in revised form 25 February 2017 was to unravel the novel role of the low-density lipoprotein receptor-related protein 3 (LRP3) in regulating Accepted 1 March 2017 the osteogenic and adipogenic differentiation of immortalized hBMSCs. Gene expression profiling revealed sig- Available online 8 March 2017 nificantly higher LRP3 levels in the highly osteogenic hBMSC clone imCL1 than in the less osteogenic clone imCL2, as well as a significant upregulation of LRP3 during the osteogenic induction of the imCL1 clone.