Phylogeography and Population Genetics of a Headwater-Stream Adapted Crayfish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Species Status Assessment Report for the Barrens Darter (Etheostoma Forbesi)
Species Status Assessment Report for the Barrens Darter (Etheostoma forbesi) Version 2.0 Acknowledgements: This Species Status Assessment would not have been possible without the research and assistance of Dr. Richard Harrington, Yale University Department of Ecology and Evolutionary Biology, Dr. Hayden Mattingly and his students, Tennessee Tech University School of Environmental Studies, Dr. John Johansen, Austin Peay State University Department of Biology, and Mark Thurman, Tennessee Wildlife Resources Agency. 1 TABLE OF CONTENTS Chapter 1: Introduction ............................................................................................................... 3 Chapter 2: Biology and Life History ........................................................................................... 4 Taxonomy ................................................................................................................................ 4 Genetic Diversity ..................................................................................................................... 5 Morphological Description ...................................................................................................... 5 Habitat ..................................................................................................................................... 6 Lifecycle .................................................................................................................................. 7 Population Needs .................................................................................................................... -

Fish Inventory at Stones River National Battlefield
Fish Inventory at Stones River National Battlefield Submitted to: Department of the Interior National Park Service Cumberland Piedmont Network By Dennis Mullen Professor of Biology Department of Biology Middle Tennessee State University Murfreesboro, TN 37132 September 2006 Striped Shiner (Luxilus chrysocephalus) – nuptial male From Lytle Creek at Fortress Rosecrans Photograph by D. Mullen Table of Contents List of Tables……………………………………………………………………….iii List of Figures………………………………………………………………………iv List of Appendices…………………………………………………………………..v Executive Summary…………………………………………………………………1 Introduction…………………………………………………………………...……..2 Methods……………………………………………………………………………...3 Results……………………………………………………………………………….7 Discussion………………………………………………………………………….10 Conclusions………………………………………………………………………...14 Literature Cited…………………………………………………………………….15 ii List of Tables Table1: Location and physical characteristics (during September 2006, and only for the riverine sites) of sample sites for the STRI fish inventory………………………………17 Table 2: Biotic Integrity classes used in assessing fish communities along with general descriptions of their attributes (Karr et al. 1986) ………………………………………18 Table 3: List of fishes potentially occurring in aquatic habitats in and around Stones River National Battlefield………………………………………………………………..19 Table 4: Fish species list (by site) of aquatic habitats at STRI (October 2004 – August 2006). MF = McFadden’s Ford, KP = King Pond, RB = Redoubt Brannan, UP = Unnamed Pond at Redoubt Brannan, LC = Lytle Creek at Fortress Rosecrans……...….22 Table 5: Fish Species Richness estimates for the 3 riverine reaches of STRI and a composite estimate for STRI as a whole…………………………………………………24 Table 6: Index of Biotic Integrity (IBI) scores for three stream reaches at Stones River National Battlefield during August 2005………………………………………………...25 Table 7: Temperature and water chemistry of four of the STRI sample sites for each sampling date…………………………………………………………………………….26 Table 8 : Total length estimates of specific habitat types at each riverine sample site. -

Evolutionary History of Inversions in the Direction of Architecture-Driven
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.09.085712; this version posted May 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Evolutionary history of inversions in the direction of architecture- driven mutational pressures in crustacean mitochondrial genomes Dong Zhang1,2, Hong Zou1, Jin Zhang3, Gui-Tang Wang1,2*, Ivan Jakovlić3* 1 Key Laboratory of Aquaculture Disease Control, Ministry of Agriculture, and State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China. 2 University of Chinese Academy of Sciences, Beijing 100049, China 3 Bio-Transduction Lab, Wuhan 430075, China * Corresponding authors Short title: Evolutionary history of ORI events in crustaceans Abbreviations: CR: control region, RO: replication of origin, ROI: inversion of the replication of origin, D-I skew: double-inverted skew, LBA: long-branch attraction bioRxiv preprint doi: https://doi.org/10.1101/2020.05.09.085712; this version posted May 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract Inversions of the origin of replication (ORI) of mitochondrial genomes produce asymmetrical mutational pressures that can cause artefactual clustering in phylogenetic analyses. It is therefore an absolute prerequisite for all molecular evolution studies that use mitochondrial data to account for ORI events in the evolutionary history of their dataset. -

Endangered Species
FEATURE: ENDANGERED SPECIES Conservation Status of Imperiled North American Freshwater and Diadromous Fishes ABSTRACT: This is the third compilation of imperiled (i.e., endangered, threatened, vulnerable) plus extinct freshwater and diadromous fishes of North America prepared by the American Fisheries Society’s Endangered Species Committee. Since the last revision in 1989, imperilment of inland fishes has increased substantially. This list includes 700 extant taxa representing 133 genera and 36 families, a 92% increase over the 364 listed in 1989. The increase reflects the addition of distinct populations, previously non-imperiled fishes, and recently described or discovered taxa. Approximately 39% of described fish species of the continent are imperiled. There are 230 vulnerable, 190 threatened, and 280 endangered extant taxa, and 61 taxa presumed extinct or extirpated from nature. Of those that were imperiled in 1989, most (89%) are the same or worse in conservation status; only 6% have improved in status, and 5% were delisted for various reasons. Habitat degradation and nonindigenous species are the main threats to at-risk fishes, many of which are restricted to small ranges. Documenting the diversity and status of rare fishes is a critical step in identifying and implementing appropriate actions necessary for their protection and management. Howard L. Jelks, Frank McCormick, Stephen J. Walsh, Joseph S. Nelson, Noel M. Burkhead, Steven P. Platania, Salvador Contreras-Balderas, Brady A. Porter, Edmundo Díaz-Pardo, Claude B. Renaud, Dean A. Hendrickson, Juan Jacobo Schmitter-Soto, John Lyons, Eric B. Taylor, and Nicholas E. Mandrak, Melvin L. Warren, Jr. Jelks, Walsh, and Burkhead are research McCormick is a biologist with the biologists with the U.S. -

Crayfishes and Shrimps) of Arkansas with a Discussion of Their Ah Bitats Raymond W
Journal of the Arkansas Academy of Science Volume 34 Article 9 1980 Inventory of the Decapod Crustaceans (Crayfishes and Shrimps) of Arkansas with a Discussion of Their aH bitats Raymond W. Bouchard Southern Arkansas University Henry W. Robison Southern Arkansas University Follow this and additional works at: http://scholarworks.uark.edu/jaas Part of the Terrestrial and Aquatic Ecology Commons Recommended Citation Bouchard, Raymond W. and Robison, Henry W. (1980) "Inventory of the Decapod Crustaceans (Crayfishes and Shrimps) of Arkansas with a Discussion of Their aH bitats," Journal of the Arkansas Academy of Science: Vol. 34 , Article 9. Available at: http://scholarworks.uark.edu/jaas/vol34/iss1/9 This article is available for use under the Creative Commons license: Attribution-NoDerivatives 4.0 International (CC BY-ND 4.0). Users are able to read, download, copy, print, distribute, search, link to the full texts of these articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author. This Article is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Journal of the Arkansas Academy of Science by an authorized editor of ScholarWorks@UARK. For more information, please contact [email protected], [email protected]. Journal of the Arkansas Academy of Science, Vol. 34 [1980], Art. 9 AN INVENTORY OF THE DECAPOD CRUSTACEANS (CRAYFISHES AND SHRIMPS) OF ARKANSAS WITH A DISCUSSION OF THEIR HABITATS i RAYMOND W. BOUCHARD 7500 Seaview Avenue, Wildwood Crest, New Jersey 08260 HENRY W. ROBISON Department of Biological Sciences Southern Arkansas University, Magnolia, Arkansas 71753 ABSTRACT The freshwater decapod crustaceans of Arkansas presently consist of two species of shrimps and 51 taxa of crayfishes divided into 47 species and four subspecies. -

A List of Common and Scientific Names of Fishes from the United States And
t a AMERICAN FISHERIES SOCIETY QL 614 .A43 V.2 .A 4-3 AMERICAN FISHERIES SOCIETY Special Publication No. 2 A List of Common and Scientific Names of Fishes -^ ru from the United States m CD and Canada (SECOND EDITION) A/^Ssrf>* '-^\ —---^ Report of the Committee on Names of Fishes, Presented at the Ei^ty-ninth Annual Meeting, Clearwater, Florida, September 16-18, 1959 Reeve M. Bailey, Chairman Ernest A. Lachner, C. C. Lindsey, C. Richard Robins Phil M. Roedel, W. B. Scott, Loren P. Woods Ann Arbor, Michigan • 1960 Copies of this publication may be purchased for $1.00 each (paper cover) or $2.00 (cloth cover). Orders, accompanied by remittance payable to the American Fisheries Society, should be addressed to E. A. Seaman, Secretary-Treasurer, American Fisheries Society, Box 483, McLean, Virginia. Copyright 1960 American Fisheries Society Printed by Waverly Press, Inc. Baltimore, Maryland lutroduction This second list of the names of fishes of The shore fishes from Greenland, eastern the United States and Canada is not sim- Canada and the United States, and the ply a reprinting with corrections, but con- northern Gulf of Mexico to the mouth of stitutes a major revision and enlargement. the Rio Grande are included, but those The earlier list, published in 1948 as Special from Iceland, Bermuda, the Bahamas, Cuba Publication No. 1 of the American Fisheries and the other West Indian islands, and Society, has been widely used and has Mexico are excluded unless they occur also contributed substantially toward its goal of in the region covered. In the Pacific, the achieving uniformity and avoiding confusion area treated includes that part of the conti- in nomenclature. -

SS-7039 (June 2016) RDA 1693 1 Department of State
Department of State For Department of State Use Only Division of Publications 312 Rosa L. Parks Avenue, 8th Floor Snodgrass/TN Tower Sequence Number: Nashville, TN 37243 Rule ID(s): Phone: 615-741-2650 File Date: Email: [email protected] Effective Date: Rulemaking Hearing Rule(s) Filing Form Rulemaking Hearing Rules are rules filed after and as a result of a rulemaking hearing (Tenn. Code Ann. § 4-5- 205). Pursuant to Tenn. Code Ann. § 4-5-229, any new fee or fee increase promulgated by state agency rule shall take effect on July 1, following the expiration of the ninety (90) day period as provided in § 4-5-207. This section shall not apply to rules that implement new fees or fee increases that are promulgated as emergency rules pursuant to § 4-5-208(a) and to subsequent rules that make permanent such emergency rules, as amended during the rulemaking process. In addition, this section shall not apply to state agencies that did not, during the preceding two (2) fiscal years, collect fees in an amount sufficient to pay the cost of operating the board, commission or entity in accordance with § 4-29-121(b). Agency/Board/Commission: Tennessee Wildlife Resources Agency Division: Biodiversity Contact Person: Lisa Crawford Address: PO Box 40747, Nashville, TN Zip: 37204 Phone: 615-781-6606 Email: [email protected] Revision Type (check all that apply): Amendment X New Repeal Rule(s) (ALL chapters and rules contained in filing must be listed here. If needed, copy and paste additional tables to accommodate multiple chapters. -
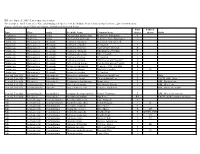
Threatened and Endangered Species List
Effective April 15, 2009 - List is subject to revision For a complete list of Tennessee's Rare and Endangered Species, visit the Natural Areas website at http://tennessee.gov/environment/na/ Aquatic and Semi-aquatic Plants and Aquatic Animals with Protected Status State Federal Type Class Order Scientific Name Common Name Status Status Habit Amphibian Amphibia Anura Gyrinophilus gulolineatus Berry Cave Salamander T Amphibian Amphibia Anura Gyrinophilus palleucus Tennessee Cave Salamander T Crustacean Malacostraca Decapoda Cambarus bouchardi Big South Fork Crayfish E Crustacean Malacostraca Decapoda Cambarus cymatilis A Crayfish E Crustacean Malacostraca Decapoda Cambarus deweesae Valley Flame Crayfish E Crustacean Malacostraca Decapoda Cambarus extraneus Chickamauga Crayfish T Crustacean Malacostraca Decapoda Cambarus obeyensis Obey Crayfish T Crustacean Malacostraca Decapoda Cambarus pristinus A Crayfish E Crustacean Malacostraca Decapoda Cambarus williami "Brawley's Fork Crayfish" E Crustacean Malacostraca Decapoda Fallicambarus hortoni Hatchie Burrowing Crayfish E Crustacean Malocostraca Decapoda Orconectes incomptus Tennessee Cave Crayfish E Crustacean Malocostraca Decapoda Orconectes shoupi Nashville Crayfish E LE Crustacean Malocostraca Decapoda Orconectes wrighti A Crayfish E Fern and Fern Ally Filicopsida Polypodiales Dryopteris carthusiana Spinulose Shield Fern T Bogs Fern and Fern Ally Filicopsida Polypodiales Dryopteris cristata Crested Shield-Fern T FACW, OBL, Bogs Fern and Fern Ally Filicopsida Polypodiales Trichomanes boschianum -

At-Risk Species Assessment on Southern National Forests, Refuges, and Other Protected Areas
David Moynahan | St. Marks NWR At-Risk Species Assessment on Southern National Forests, Refuges, and Other Protected Areas National Wildlife Refuge Association Mark Sowers, Editor October 2017 1001 Connecticut Avenue NW, Suite 905, Washington, DC 20036 • 202-417-3803 • www.refugeassociation.org At-Risk Species Assessment on Southern National Forests, Refuges, and Other Protected Areas Table of Contents Introduction and Methods ................................................................................................3 Results and Discussion ......................................................................................................9 Suites of Species: Occurrences and Habitat Management ...........................................12 Progress and Next Steps .................................................................................................13 Appendix I: Suites of Species ..........................................................................................17 Florida Panhandle ............................................................................................................................18 Peninsular Florida .............................................................................................................................28 Southern Blue Ridge and Southern Ridge and Valley ...............................................................................................................................39 Interior Low Plateau and Cumberland Plateau, Central Ridge and Valley ...............................................................................................46 -

Synopsis of Freshwater Crayfish Diseases and Commensal Organisms Brett .F Edgerton James Cook University, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications from the Harold W. Manter Parasitology, Harold W. Manter Laboratory of Laboratory of Parasitology 3-2002 Synopsis of Freshwater Crayfish Diseases and Commensal Organisms Brett .F Edgerton James Cook University, [email protected] Louis H. Evans Curtin University of Technology Frances J. Stephens Curtin University of Technology Robin M. Overstreet Gulf Coast Research Laboratory, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/parasitologyfacpubs Part of the Aquaculture and Fisheries Commons, and the Parasitology Commons Edgerton, Brett .;F Evans, Louis H.; Stephens, Frances J.; and Overstreet, Robin M., "Synopsis of Freshwater Crayfish Diseases and Commensal Organisms" (2002). Faculty Publications from the Harold W. Manter Laboratory of Parasitology. 884. https://digitalcommons.unl.edu/parasitologyfacpubs/884 This Article is brought to you for free and open access by the Parasitology, Harold W. Manter Laboratory of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications from the Harold W. Manter Laboratory of Parasitology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published in Aquaculture 206:1–2 (March 2002), pp. 57–135; doi: 10.1016/S0044-8486(01)00865-1 Copyright © 2002 Elsevier Science. Creative Commons Attribution Non-Commercial No Deriva- tives License. Accepted October 18, 2001; published online November 30, 2001. Synopsis of Freshwater Crayfish Diseases and Commensal Organisms Brett F. Edgerton,1 Louis H. Evans,2 Frances J. Stephens,2 and Robin M. Overstreet3 1. Department of Microbiology and Immunology, James Cook University, Townsville, QLD 4810, Australia 2. -

Endangered Species
FEATURE: ENDANGERED SPECIES Conservation Status of Imperiled North American Freshwater and Diadromous Fishes ABSTRACT: This is the third compilation of imperiled (i.e., endangered, threatened, vulnerable) plus extinct freshwater and diadromous fishes of North America prepared by the American Fisheries Society’s Endangered Species Committee. Since the last revision in 1989, imperilment of inland fishes has increased substantially. This list includes 700 extant taxa representing 133 genera and 36 families, a 92% increase over the 364 listed in 1989. The increase reflects the addition of distinct populations, previously non-imperiled fishes, and recently described or discovered taxa. Approximately 39% of described fish species of the continent are imperiled. There are 230 vulnerable, 190 threatened, and 280 endangered extant taxa, and 61 taxa presumed extinct or extirpated from nature. Of those that were imperiled in 1989, most (89%) are the same or worse in conservation status; only 6% have improved in status, and 5% were delisted for various reasons. Habitat degradation and nonindigenous species are the main threats to at-risk fishes, many of which are restricted to small ranges. Documenting the diversity and status of rare fishes is a critical step in identifying and implementing appropriate actions necessary for their protection and management. Howard L. Jelks, Frank McCormick, Stephen J. Walsh, Joseph S. Nelson, Noel M. Burkhead, Steven P. Platania, Salvador Contreras-Balderas, Brady A. Porter, Edmundo Díaz-Pardo, Claude B. Renaud, Dean A. Hendrickson, Juan Jacobo Schmitter-Soto, John Lyons, Eric B. Taylor, and Nicholas E. Mandrak, Melvin L. Warren, Jr. Jelks, Walsh, and Burkhead are research McCormick is a biologist with the biologists with the U.S. -

A Synthetic Phylogeny of Freshwater Crayfish: Insights for Conservation Rstb.Royalsocietypublishing.Org Christopher L
Downloaded from http://rstb.royalsocietypublishing.org/ on January 27, 2015 A synthetic phylogeny of freshwater crayfish: insights for conservation rstb.royalsocietypublishing.org Christopher L. Owen1, Heather Bracken-Grissom2, David Stern1 and Keith A. Crandall1,3 1Computational Biology Institute, George Washington University, Ashburn, VA 20132, USA Research 2Department of Biological Sciences, Florida International University, Biscayne Bay Campus, North Miami, FL 33181, USA 3 Cite this article: Owen CL, Bracken-Grissom Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, DC 20013, USA H, Stern D, Crandall KA. 2015 A synthetic phylogeny of freshwater crayfish: insights for Phylogenetic systematics is heading for a renaissance where we shift from conservation. Phil. Trans. R. Soc. B 370: considering our phylogenetic estimates as a static image in a published 20140009. paper and taxonomies as a hardcopy checklist to treating both the phyloge- http://dx.doi.org/10.1098/rstb.2014.0009 netic estimate and dynamic taxonomies as metadata for further analyses. The Open Tree of Life project (opentreeoflife.org) is developing synthesis tools for harnessing the power of phylogenetic inference and robust taxonomy One contribution of 17 to a discussion meeting to develop a synthetic tree of life. We capitalize on this approach to estimate a issue ‘Phylogeny, extinction and conservation’. synthesis tree for the freshwater crayfish. The crayfish make an exceptional group to demonstrate the utility of the synthesis approach, as there recently Subject Areas: have been a number of phylogenetic studies on the crayfishes along with a robust underlying taxonomic framework. Importantly, the crayfish have also evolution, taxonomy and systematics, been extensively assessed by an IUCN Red List team and therefore have accu- environmental science rate and up-to-date area and conservation status data available for analysis within a phylogenetic context.