Blueprint Genetics Progeria and Progeroid Syndromes Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New ZMPSTE24 (FACE1) Mutations in Patients Affected with Restrictive
New ZMPSTE24 (FACE1) mutations in patients affected with restrictive dermopathy or related progeroid syndromes and mutation update Claire Navarro, Patrice Roll, Vera Esteves-Vieira, Sebastien Courrier, Amandine Boyer, Thuy Duong Nguyen, Le Thi Thanh Huong, Peter Meinke, Winnie Schröder, Valérie Cormier-Daire, et al. To cite this version: Claire Navarro, Patrice Roll, Vera Esteves-Vieira, Sebastien Courrier, Amandine Boyer, et al.. New ZMPSTE24 (FACE1) mutations in patients affected with restrictive dermopathy or related progeroid syndromes and mutation update. European Journal of Human Genetics, Nature Publishing Group, 2013, 22 (8), pp.1002-1011. 10.1038/ejhg.2013.258. hal-01664301 HAL Id: hal-01664301 https://hal-amu.archives-ouvertes.fr/hal-01664301 Submitted on 20 Dec 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. New ZMPSTE24 (FACE1) mutations in patients affected with restrictive dermopathy or related progeroid syndromes and mutation update Claire Laure Navarro*,1,2, Vera Esteves-Vieira3,Se´bastien Courrier1,2, Amandine Boyer3, Thuy Duong -

De Barsy Syndrome: Orthopedic Case Report and Literature Review
MOJ Orthopedics & Rheumatology De Barsy Syndrome: Orthopedic Case report and Literature Review Introduction Case Report Volume 7 Issue 5 - 2017 This condition was first described in 1968 by De Barsy who and degeneration of the elastic tissue of the cornea and skin, Jose de Jesus Guerra Jasso1*, Douglas reported a case of a patient with progeria, dwarfism, oligofrenia and since then, it is known as Barsy Syndrome or Barsy- Colmenares Bonilla2 and Loreett Ocampo Perez3 of progeroid aspect, cutis laxa, corneal opacity, intrauterine 1Pediatric Orthopedics, Hospital Regional de Alta Especialidad growthMoens-Dierckx retardation Syndrome. and severe This ismental defined retardation as the combination (although del Bajio, Mexico some will learn to speak, intelligence is less than normal) [1]. 2Pediatric Orthopedic Service, Hospital regional de alta especialidad del bajio The orthopedic manifestations are dysplasia of hip development, 3Fellow of Pediatric Orthopedics, Hospital Regional de Alta hyper laxity of severe joints, athetoid movements, scoliosis and Especialidad del Bajio, Mexico severe deformities of the foot. Epidemiology in Latin America is unknown because of its underdiagnosis and when confused *Corresponding author: Jesus Guerra Jasso, Hospital with other connective tissue pathologies (Hutchinson-Gilford Regional de Alta Especialidad del Bajio, Boulevard Milenio No. 130. Col, San Carlos la Roncha, C.P. 37660, Guanajuato, syndrome, gerodermic osteodiplasia, even Ehlers-Danlos), the Mexico, Tel: 477-267 2000; Ext-1403; life expectancy of these patients varies according to the degree of Email: penetrance and in the world literature, there are very few reports (about 50), so the diagnosis requires a challenge [2]. Received: January 13, 2017 | Published: March 21, 2017 Clinical Case and thick clamp. -

Open Full Page
CCR PEDIATRIC ONCOLOGY SERIES CCR Pediatric Oncology Series Recommendations for Childhood Cancer Screening and Surveillance in DNA Repair Disorders Michael F. Walsh1, Vivian Y. Chang2, Wendy K. Kohlmann3, Hamish S. Scott4, Christopher Cunniff5, Franck Bourdeaut6, Jan J. Molenaar7, Christopher C. Porter8, John T. Sandlund9, Sharon E. Plon10, Lisa L. Wang10, and Sharon A. Savage11 Abstract DNA repair syndromes are heterogeneous disorders caused by around the world to discuss and develop cancer surveillance pathogenic variants in genes encoding proteins key in DNA guidelines for children with cancer-prone disorders. Herein, replication and/or the cellular response to DNA damage. The we focus on the more common of the rare DNA repair dis- majority of these syndromes are inherited in an autosomal- orders: ataxia telangiectasia, Bloom syndrome, Fanconi ane- recessive manner, but autosomal-dominant and X-linked reces- mia, dyskeratosis congenita, Nijmegen breakage syndrome, sive disorders also exist. The clinical features of patients with DNA Rothmund–Thomson syndrome, and Xeroderma pigmento- repair syndromes are highly varied and dependent on the under- sum. Dedicated syndrome registries and a combination of lying genetic cause. Notably, all patients have elevated risks of basic science and clinical research have led to important in- syndrome-associated cancers, and many of these cancers present sights into the underlying biology of these disorders. Given the in childhood. Although it is clear that the risk of cancer is rarity of these disorders, it is recommended that centralized increased, there are limited data defining the true incidence of centers of excellence be involved directly or through consulta- cancer and almost no evidence-based approaches to cancer tion in caring for patients with heritable DNA repair syn- surveillance in patients with DNA repair disorders. -

DNA Damage in the Oligodendrocyte Lineage and Its Role in Brain Aging
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Mech Ageing Manuscript Author Dev. Author Manuscript Author manuscript; available in PMC 2018 January 01. Published in final edited form as: Mech Ageing Dev. 2017 January ; 161(Pt A): 37–50. doi:10.1016/j.mad.2016.05.006. DNA damage in the oligodendrocyte lineage and its role in brain aging Kai-Hei Tse1,2 and Karl Herrup1 1 Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong Abstract Myelination is a recent evolutionary addition that significantly enhances the speed of transmission in the neural network. Even slight defects in myelin integrity impair performance and enhance the risk of neurological disorders. Indeed, myelin degeneration is an early and well-recognized neuropathology that is age associated, but appears before cognitive decline. Myelin is only formed by fully differentiated oligodendrocytes, but the entire oligodendrocyte lineage are clear targets of the altered chemistry of the aging brain. As in neurons, unrepaired DNA damage accumulates in the postmitotic oligodendrocyte genome during normal aging, and indeed may be one of the upstream causes of cellular aging - a fact well illustrated by myelin co-morbidity in premature aging syndromes arising from deficits in DNA repair enzymes. The clinical and experimental evidence from Alzheimer's disease, progeroid syndromes, ataxia-telangiectasia and other conditions strongly suggest that oligodendrocytes may in fact be uniquely vulnerable to oxidative DNA damage. If this damage remains unrepaired, as is increasingly true in the aging brain, myelin gene transcription and oligodendrocyte differentiation is impaired. Delineating the relationships between early myelin loss and DNA damage in brain aging will offer an additional dimension outside the neurocentric view of neurodegenerative disease. -

The Progeria Syndrome Fact Sheet
HUTCHINSON-GILFORD PROGERIA SYNDROME FREQUENTLY ASKED QUESTIONS WHAT IS PROGERIA? Hutchinson-Gilford Progeria Syndrome “Progeria” or “HGPS” is a rare, fatal genetic condition characterized by an appearance of accelerated aging in children. Its name is derived from the Greek and means "prematurely old." While there are different forms of Progeria*, the classic type is Hutchinson- Gilford Progeria Syndrome, which was named after the doctors who first described it in England: in 1886 by Dr. Jonathan Hutchinson, and in 1897 by Dr. Hastings Gilford. HOW COMMON IS PROGERIA? Progeria affects approximately 1 in 4 - 8 million newborns. It affects both sexes equally and all races. In the past 15 years, children with Progeria have been reported all over the world , including in: Algeria Cuba Ireland Peru Sweden Argentina Denmark Israel Philippines Switzerland Australia Dominican Italy Poland Turkey Austria Republic Japan Portugal United States Belgium Egypt Libya Puerto Rico Venezuela Brazil England Mexico Romania Vietnam Canada France Morocco South Africa Yugoslavia China Germany Netherlands South Korea Columbia India Pakistan Spain WHAT ARE THE FEATURES OF PROGERIA? Although they are born looking healthy, most children with Progeria begin to display many characteristics of Progeria within the first year of life. Progeria signs include growth failure, loss of body fat and hair, aged-looking skin, stiffness of joints, hip dislocation, generalized atherosclerosis, cardiovascular (heart) disease and stroke. The children have a remarkably similar appearance, despite differing ethnic backgrounds. Children with Progeria die of atherosclerosis (heart disease) at an average age of thirteen years (with a range of about 8 – 21 years). WHAT DOES PROGERIA HAVE TO DO WITH AGING? Children with Progeria are genetically predisposed to premature, progressive heart disease. -

Atypical Progeroid Syndrome ID: 20-0188; April 2021 (P.E262K LMNA) DOI: 10.1530/EDM-20-0188
ID: 20-0188 -20-0188 M Yukina and others Atypical progeroid syndrome ID: 20-0188; April 2021 (p.E262K LMNA) DOI: 10.1530/EDM-20-0188 Atypical progeroid syndrome (p.E262K LMNA mutation): a rare cause of short stature and osteoporosis Correspondence Marina Yukina, Nurana Nuralieva, Ekaterina Sorkina, Ekaterina Troshina, should be addressed Anatoly Tiulpakov, Zhanna Belaya and Galina Melnichenko to E Sorkina Email Endocrinology Research Centre, Moscow, Russia [email protected] Summary Lamin A/C (LMNA) gene mutations cause a heterogeneous group of progeroid disorders, including Hutchinson–Gilford progeria syndrome, mandibuloacral dysplasia, atypical progeroid syndrome (APS) and generalized lipodystrophy- associated progeroid syndrome (GLPS). All of those syndromes are associated with some progeroid features, lipodystrophyandmetaboliccomplicationsbutvarydifferentlydependingonaparticularmutationandevenpatients carrying the same gene variant are known to have clinical heterogeneity. We report a new 30-year-old female patient from Russia with an APS and generalized lipodystrophy (GL) due to the heterozygous de novo LMNA p.E262K mutation and compare her clinical and metabolic features to those of other described patients with APS. Despite many health issues, short stature, skeletal problems, GL and late diagnosis of APS, our patient seems to be relatively metabolically healthy for her age when compared to previously described patients with APS. Learning points: • Atypicalprogeroidsyndromes(APS)arerareandheterogenicwithdifferentageofonsetanddegreeofmetabolic -
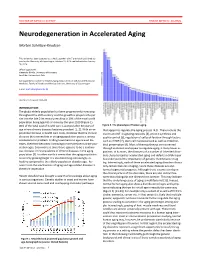
Neurodegeneration in Accelerated Aging
DOCTOR OF MEDICAL SCIENCE DANISH MEDICAL JOURNAL Neurodegeneration in Accelerated Aging Morten Scheibye-Knudsen This review has been accepted as a thesis together with 7 previously published pa- pers by the University of Copenhagen, October 16, 2014 and defended on January 14, 2016 Official opponents: Alexander Bürkle, University of Konstanz Lars Eide, University of Oslo Correspondence: Center for Healthy Aging, Department of Cellular and Molecular Medicine, Faculty of Health and Medical Sciences, University of Copenhagen E-mail: [email protected] Dan Med J 2016;63(11):B5308 INTRODUCTION The global elderly population has been progressively increasing throughout the 20th century and this growth is projected to per- sist into the late 21st century resulting in 20% of the total world population being aged 65 or more by the year 2100 (Figure 1). 80% of the total cost of health care is accrued after 40 years of Figure 2. The phenotype of human aging. age where chronic diseases become prevalent [1, 2]. With an ex- that appear to regulate the aging process [4,5]. These include the ponential increase in health care costs, it follows that the chronic insulin and IGF-1 signaling cascades [4], protein synthesis and diseases that accumulate in an aging population poses a serious quality control [6], regulation of cell proliferation through factors socioeconomic problem. Finding treatments to age related dis- such as mTOR [7], stem cell maintenance 8 as well as mitochon- eases, therefore becomes increasingly more pertinent as the pop- drial preservation [9]. Most of these pathways are conserved ulation ages. Even more so since there appears to be a continu- through evolution and appear to regulate aging in many lower or- ous increase in the prevalence of chronic diseases in the aging ganisms. -
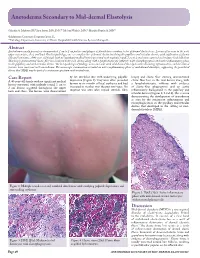
Anetoderma Secondary to Mid-Dermal Elastolysis
Anetoderma Secondary to Mid-dermal Elastolysis Gabriela A. Maloney, BS,* Jane James, MD, PhD,** Michael Welsch, MD,** Marylee Braniecki, MD** *Midwestern University, Downers Grove, IL **Pathology Department, University of Illinois Hospital & Health Sciences System, Chicago, IL Abstract Anetoderma usually presents as circumscribed, 1 cm to 2 cm patches and plaques of flaccid skin secondary to loss of dermal elastic tissue. Lesions often occur in the neck, upper extremities, chest, and back. On histopathology, one sees complete loss of dermal elastin involving the papillary and reticular dermis, with infiltration of plasma cells and histiocytes. A 40-year-old female with no significant medical history presented with multiple round, 1 cm to 2 cm lesions scattered on her upper back and chest. Skin biopsy demonstrated elastic-fiber loss localized to the mid-dermis along with a lymphohistiocytic infiltrate with elastophagocytosis and active inflammatory phase in the papillary and mid-reticular dermis. The histopathological findings were consistent with mid-dermal elastolysis with advancing inflammation, and the clinical features were consistent with anetoderma. The microscopic examination revealed an active inflammatory phase of mid-dermal elastolysis, supporting the postulated theory that MDE may be part of a continuous spectrum with anetoderma. Case Report by lax, wrinkled skin with underlying palpable biopsy and elastic-fiber staining demonstrated A 40-year-old female with no significant medical depression (Figure 1). They were often preceded elastic-fiber loss in the mid-dermis along with history presented with multiple round, 1 cm to by two to six months of local erythema and had a lymphohistiocytic infiltrate with evidence 2 cm lesions scattered throughout the upper increased in number over the past two years. -

Trichothiodystrophy
Trichothiodystrophy Author: Doctor Alfredo Rossi1 and Doctor C. Cantisani. Creation date: June 2004 Scientific Editor: Prof Antonella Tosti 1Dipartimento di Malattie Cutanee-Veneree Chirurgia Plastica-Ricostruttiva, Università degli studi di Roma “La Sapienza” Abstract Keywords Definition Epidemiology Etiology Clinical description Diagnostic methods Prenatal diagnosis Management References Abstract Trichothiodystrophy (TTD) is a rare autosomal recessive genetic disorder characterized by abnormal synthesis of the sulphur containing keratins and consequently hair dysplasia, associated with numerous symptoms affecting mainly organs derived from the neuroectoderm. This phenotypic aspect is due to mutations in the DNA-dependent ATPase/helicase subunit of TFIIH, XPB and XPD. Abnormalities in excision repair of ultraviolet (UV)-damaged DNA are recognized in about half of the patients. The clinical appearance is characterized by brittle and fragile hair, congenital ichthyosis, nail and dental dysplasias, cataract, progeria-like face, growth and mental retardation. The abnormalities are usually obvious at birth, with variable clinical expression. The variants of TTD, depending on their different associations, are known by the initials BIDS, IBIDS, PIBIDS, SIBIDS, ONMRS, as well as the eponyms of the Pollit, Tay, Sabinas syndromes or Amish brittle hair. The exact prevalence of TTD is unknown, but appears to be rather uncommon. About 20 cases of PIBI(D)S have been reported in the literature. Up to 1991, clinical data of 15 cases with IBIDS were published. Prenatal diagnostic of TTD is available. There is no specific treatment. Keywords Brittle hair, photosensitivity, ichthyosis, BIDS, IBIDS, PIBIDS, SIBIDS, ONMRS, Tay-syndrome Definition tail pattern). They named it Trichothiodystrophy, Trichothiodystrophy (TTD) is a group of rare noticing also an increased Photosensitivity and autosomal recessive disorders with heterogenic Ichthyosis in these patients (PIBIDS). -

De Barsy Syndrome
orphananesthesia Anesthesia recommendations for patients suffering from De Barsy syndrome Disease name: De Barsy syndrome ICD 10: Q87.7; OMIM 614438 Synonyms: DBS, De Barsy-Moens-Dierckx syndrome, Progeroid syndrome of De Barsy, Autosomal recessive cutis laxa Type 3 *With 2 gene subdivisions: ARCL3A: caused by a ALDH18A1 mutation ARCL3B: caused by a PYCR1 mutation DeBarsy syndrome is a rare clinical syndrome characterized by cutis laxa, ophthalmic opacification, skeletal malformations, as well as mental and growth retardation. This disease is genetically transmitted in an autosomal recessive fashion. Affected patients often require surgical correction of ophthalmic and orthopaedic abnormalities. This syndrome was first described by A.M. De Barsy in 1967 and less than 100 known cases are documented in the medical literature. Very little has been published on this rare disorder and only a single article has addressed anesthesia case outcomes and management strategies (Aponte, 2010). The diverse collection of clinical manifestations in De Barsy syndrome includes: intra-uterine growth retardation (IUGR), postnatal growth delay, motor delay, cognitive impairment, hypontonia, athetoid movements, malformations, microcephaly, wormian bones, large fontanelles, facial dysmorphism, cataracts, corneal clouding, thin/wrinkled skin, easy bruising, sparse hair, joint laxity, osteopenia, and inguinal hernias. Medicine in progress Perhaps new knowledge Every patient is unique Perhaps the diagnostic is wrong Find more information on the disease, its centres -

1. Progeria 101: Frequently Asked Questions
1. PROGERIA 101: FREQUENTLY ASKED QUESTIONS 1. Progeria 101: Frequently Asked Questions What is Hutchinson-Gilford Progeria Syndrome? What is PRF’s history and mission? What causes Progeria? How is Progeria diagnosed? Are there different types of Progeria? Is Progeria contagious or inherited? What is Hutchinson-Gilford Progeria Syndrome (HGPS or Progeria)? Progeria is also known as Hutchinson-Gilford Progeria Syndrome (HGPS). Genetic testing for It was first described in 1886 by Dr. Jonathan Hutchinson and in 1897 by Progeria can be Dr. Hastings Gilford. Progeria is a rare, fatal, “premature aging” syndrome. It’s called a syndrome performed from a because all the children have very similar symptoms that “go together”. The small sample of blood children have a remarkably similar appearance, even though Progeria affects children of all different ethnic backgrounds. Although most babies with (1-2 tsp) or sometimes Progeria are born looking healthy, they begin to display many characteristics from a sample of saliva. of accelerated aging by 18-24 months of age, or even earlier. Progeria signs include growth failure, loss of body fat and hair, skin changes, stiffness of joints, hip dislocation, generalized atherosclerosis, cardiovascular (heart) disease, and stroke. Children with Progeria die of atherosclerosis (heart disease) or stroke at an average age of 13 years (with a range of about 8-21 years). Remarkably, the intellect of children with Progeria is unaffected, and despite the physical changes in their young bodies, these extraordinary children are intelligent, courageous, and full of life. 1.2 THE PROGERIA HANDBOOK What is PRF’s history and mission? The Progeria Research Foundation (PRF) was established in the United States in 1999 by the parents of a child with Progeria, Drs. -

Hereditary Hearing Impairment with Cutaneous Abnormalities
G C A T T A C G G C A T genes Review Hereditary Hearing Impairment with Cutaneous Abnormalities Tung-Lin Lee 1 , Pei-Hsuan Lin 2,3, Pei-Lung Chen 3,4,5,6 , Jin-Bon Hong 4,7,* and Chen-Chi Wu 2,3,5,8,* 1 Department of Medical Education, National Taiwan University Hospital, Taipei City 100, Taiwan; [email protected] 2 Department of Otolaryngology, National Taiwan University Hospital, Taipei 11556, Taiwan; [email protected] 3 Graduate Institute of Clinical Medicine, National Taiwan University College of Medicine, Taipei City 100, Taiwan; [email protected] 4 Graduate Institute of Medical Genomics and Proteomics, National Taiwan University College of Medicine, Taipei City 100, Taiwan 5 Department of Medical Genetics, National Taiwan University Hospital, Taipei 10041, Taiwan 6 Department of Internal Medicine, National Taiwan University Hospital, Taipei 10041, Taiwan 7 Department of Dermatology, National Taiwan University Hospital, Taipei City 100, Taiwan 8 Department of Medical Research, National Taiwan University Biomedical Park Hospital, Hsinchu City 300, Taiwan * Correspondence: [email protected] (J.-B.H.); [email protected] (C.-C.W.) Abstract: Syndromic hereditary hearing impairment (HHI) is a clinically and etiologically diverse condition that has a profound influence on affected individuals and their families. As cutaneous findings are more apparent than hearing-related symptoms to clinicians and, more importantly, to caregivers of affected infants and young individuals, establishing a correlation map of skin manifestations and their underlying genetic causes is key to early identification and diagnosis of syndromic HHI. In this article, we performed a comprehensive PubMed database search on syndromic HHI with cutaneous abnormalities, and reviewed a total of 260 relevant publications.