R in Linux Or Ios: Startup
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Answers to Even- Numbered Exercises 5
Answers to Even- Numbered Exercises 5 from page 163 1. What does the shell ordinarily do while a command is executing? What should you do if you do not want to wait for a command to finish before running another command? 2. Using sort as a filter, rewrite the following sequence of commands: $ sort list > temp $ lpr temp $ rm temp $ cat list | sort | lpr 3. What is a PID number? Why are they useful when you run processes in the background? 4. Assume that the following files are in the working directory: $ ls intro notesb ref2 section1 section3 section4b notesa ref1 ref3 section2 section4a sentrev Give commands for each of the following, using wildcards to express filenames with as few characters as possible. 1 2 Chapter 5 Answers to Exercises a. List all files that begin with section. $ ls section* b. List the section1, section2, and section3 files only. $ ls section[1-3] c. List the intro file only. $ ls i* d. List the section1, section3, ref1, and ref3 files. $ ls *[13] 5. Refer to the documentation of utilities in Part III or the man pages to determine what commands will a. Output the number of lines in the standard input that contain the word a or A. b. Output only the names of the files in the working directory that contain the pattern $(. c. List the files in the working directory in their reverse alphabetical order. d. Send a list of files in the working directory to the printer, sorted by size. 6. Give a command to a. Redirect the standard output from a sort command into a file named phone_list. -

Your Performance Task Summary Explanation
Lab Report: 11.2.5 Manage Files Your Performance Your Score: 0 of 3 (0%) Pass Status: Not Passed Elapsed Time: 6 seconds Required Score: 100% Task Summary Actions you were required to perform: In Compress the D:\Graphics folderHide Details Set the Compressed attribute Apply the changes to all folders and files In Hide the D:\Finances folder In Set Read-only on filesHide Details Set read-only on 2017report.xlsx Set read-only on 2018report.xlsx Do not set read-only for the 2019report.xlsx file Explanation In this lab, your task is to complete the following: Compress the D:\Graphics folder and all of its contents. Hide the D:\Finances folder. Make the following files Read-only: D:\Finances\2017report.xlsx D:\Finances\2018report.xlsx Complete this lab as follows: 1. Compress a folder as follows: a. From the taskbar, open File Explorer. b. Maximize the window for easier viewing. c. In the left pane, expand This PC. d. Select Data (D:). e. Right-click Graphics and select Properties. f. On the General tab, select Advanced. g. Select Compress contents to save disk space. h. Click OK. i. Click OK. j. Make sure Apply changes to this folder, subfolders and files is selected. k. Click OK. 2. Hide a folder as follows: a. Right-click Finances and select Properties. b. Select Hidden. c. Click OK. 3. Set files to Read-only as follows: a. Double-click Finances to view its contents. b. Right-click 2017report.xlsx and select Properties. c. Select Read-only. d. Click OK. e. -

What Is UNIX? the Directory Structure Basic Commands Find
What is UNIX? UNIX is an operating system like Windows on our computers. By operating system, we mean the suite of programs which make the computer work. It is a stable, multi-user, multi-tasking system for servers, desktops and laptops. The Directory Structure All the files are grouped together in the directory structure. The file-system is arranged in a hierarchical structure, like an inverted tree. The top of the hierarchy is traditionally called root (written as a slash / ) Basic commands When you first login, your current working directory is your home directory. In UNIX (.) means the current directory and (..) means the parent of the current directory. find command The find command is used to locate files on a Unix or Linux system. find will search any set of directories you specify for files that match the supplied search criteria. The syntax looks like this: find where-to-look criteria what-to-do All arguments to find are optional, and there are defaults for all parts. where-to-look defaults to . (that is, the current working directory), criteria defaults to none (that is, select all files), and what-to-do (known as the find action) defaults to ‑print (that is, display the names of found files to standard output). Examples: find . –name *.txt (finds all the files ending with txt in current directory and subdirectories) find . -mtime 1 (find all the files modified exact 1 day) find . -mtime -1 (find all the files modified less than 1 day) find . -mtime +1 (find all the files modified more than 1 day) find . -

List of MS-DOS Commands - Wikipedia, the Free Encyclopedia Page 1 of 25
List of MS-DOS commands - Wikipedia, the free encyclopedia Page 1 of 25 List of MS-DOS commands From Wikipedia, the free encyclopedia In the personal computer operating systems MS -DOS and PC DOS, a number of standard system commands were provided for common Contents tasks such as listing files on a disk or moving files. Some commands were built-in to the command interpreter, others existed as transient ■ 1 Resident and transient commands commands loaded into memory when required. ■ 2 Command line arguments Over the several generations of MS-DOS, ■ 3 Windows command prompt commands were added for the additional ■ 4 Commands functions of the operating system. In the current ■ 4.1 @ Microsoft Windows operating system a text- ■ 4.2 : mode command prompt window can still be ■ 4.3 ; used. Some DOS commands carry out functions ■ 4.4 /* equivalent to those in a UNIX system but ■ 4.5 ( ) always with differences in details of the ■ 4.6 append function. ■ 4.7 assign ■ 4.8 attrib ■ 4.9 backup and restore Resident and transient ■ 4.10 BASIC and BASICA commands ■ 4.11 call ■ 4.12 cd or chdir ■ 4.13 chcp The command interpreter for MS-DOS runs ■ 4.14 chkdsk when no application programs are running. ■ 4.15 choice When an application exits, if the command ■ 4.16 cls interpreter in memory was overwritten, MS- ■ 4.17 copy DOS will re-load it from disk. The command ■ 4.18 ctty interpreter is usually stored in a file called ■ 4.19 defrag "COMMAND.COM". Some commands are ■ 4.20 del or erase internal and built-into COMMAND.COM, ■ 4.21 deltree others are stored on disk in the same way as ■ 4.22 dir application programs. -

L01 – the Terminal • What Is a Terminal? O Command Line Interface with the Computer. Text Only. in the Old Days the Terminal
L01 – The Terminal • What is a terminal? o Command line interface with the computer. Text only. In the old days the terminal was the only interface to the computer. o Terminal, bash, shell, console, are all interchangeable words, as far as we’re concerned. o The terminal will be our programming environment, we won’t use IDE’s like Eclipse. • Using the terminal o Everything you can do using the Finder, such as navigate to different folders, open files, create files and directories, can also be done via the terminal. o Navigating the file system § “pwd” – present working directory; reports your current location in the file system § “cd <dir>” – change directory; goes into a specified directory § “ls” – list; lists the contents of your pwd • “ls –alF” – the options “-alF” lists all files (even hidden files) with extra information. § “.” and “..” – shortcuts that refer to current directory, and up one level directory • e.g.: “cd ..” takes you up one level. “cd .” takes your to the current directory, effectively does nothing. § “~” is a shortcut that refers to your home directory. For me its /Users/gregorychen3 o Making changes to the file system § “mkdir <name>” – make directory; creates a new directory (folder) called <name> § “cp <src> <dest>” – copy; copies file located at <src> to <dest>. Hint: use “.” and “..” shortcuts to specify the file paths § “mv <src> <dest>” – move; move a file located at <src> to <dest>. Hint: use “.” and “..” shortcuts to specify the file paths § “rm <file>” – remove; removes the file located at <file>. • “rm –r <dir>” – remove recursive; removes directory located at <dir>. -

Windows Powershell Best Practices Windows Powershell Best Practices
Windows PowerShell Best Practices Windows PowerShell Best Practices Expert recommendations, pragmatically applied Automate system administration using Windows PowerShell best practices—and optimize your operational efficiency. With this About the Author practical guide, Windows PowerShell expert and instructor Ed Ed Wilson, MCSE, CISSP, is a well-known Wilson delivers field-tested tips, real-world examples, and candid scripting expert and author of “Hey Windows Scripting Guy!”—one of the most popular advice culled from administrators across a range of business and blogs on Microsoft TechNet. He’s written technical scenarios. If you’re an IT professional with Windows several books on Windows scripting PowerShell experience, this book is ideal. for Microsoft Press, including Windows PowerShell 2.0 Best Practices and Windows PowerShell Scripting Guide. Discover how to: PowerShell • Use Windows PowerShell to automate Active Directory tasks • Explore available WMI classes and methods with CIM cmdlets • Identify and track scripting opportunities to avoid duplication • Use functions to encapsulate business logic and reuse code • Design your script’s best input method and output destination • Test scripts by checking their syntax and performance • Choose the most suitable method for running remote commands • Manage software services with Desired State Configuration Wilson BEST PRACTICES microsoft.com/mspress ISBN 978-0-7356-6649-8 U.S.A. $59.99 55999 Canada $68.99 [Recommended] 9 780735 666498 Operating Systems/Windows Server Celebrating 30 years! Ed Wilson 666498_Win_PowerShell_Best_Practices.indd 1 4/11/14 10:30 AM Windows PowerShell Best Practices Ed Wilson 666498_book.indb 1 12/20/13 10:50 AM Published with the authorization of Microsoft Corporation by: O’Reilly Media, Inc. -

Introduction to the Command Line Getting Started with Windows Powershell
Introduction To The Command Line Getting started with Windows Powershell The Command Line is a text-based interface that allows you to communicate with your computer to accomplish a wide range of tasks. You will use the skills you develop in this lesson when working with Twarc, a command line tool for collecting and analyzing Twitter data. It is important to have a handle on basic commands so that you can work with the 1 October Twitter Data Collection later on in this tutorial series. Get started by downloading the materials below! Difficulty level: Beginner Optimized for: Windows users. Mac users can view the tutorial here. Prerequisite(s): None Materials: Download ‘walt_whitman.zip’ to your desktop Tutorial Key ● Command Line commands will be displayed in this format ● means you have come to the end of a set of instructions Lesson objectives - Use the command line to navigate your computer - Create and move content around - Make changes to existing files Key Terms ● PowerShell - Command Line Shell from Microsoft ○ A text interface for your computer. PowerShell receives commands and then passes those commands on to the computer's operating system to run. ● Command ○ A specific order from a user to the computer’s operating system to perform a service ● Graphical-User Interface (GUI) ○ A visual form of user interface that allows users to interact with a computer through icons and other visual indicators ● Filepath ○ A unique address that specifies a location in a file system ● Directory ○ A location for storing files on your computer. A directory is the same thing as a folder; a folder is represented visually in a GUI. -

Fundamentals of UNIX Lab 5.3.3 – Navigating the File System (Estimated Time: 20 Min.)
Fundamentals of UNIX Lab 5.3.3 – Navigating the File System (Estimated time: 20 min.) Objectives: • Become familiar with the file system directory tree used in class • Determine the current working directory with the pwd command • Work with absolute and relative pathnames • Change directories from one location to another in the file system with the cd command Background: In this lab the student will work with the UNIX file system or directory tree, which has been set up for the class. The student will learn how to determine their current location in the directory tree and how to change from one directory to another. Remembering which directory a user is currently working in is often difficult. The pwd (print working directory) command will display the absolute pathname of the current directory. The pwd command is used frequently to check the current location in the directory tree or hierarchy. A user can move from one directory to another with the cd (change directory) command. The directory location a user wishes to change to is specified using an absolute or relative pathname. An absolute pathname specifies a file or directory in relation to the entire file hierarchy. The hierarchy begins at the / (root) directory. Absolute pathnames always start at the root (/) directory and list each directory along the path to the destination file or directory. A relative pathname describes the location of a file or directory as it relates to the current directory or the directory a user is currently in. If a user is in a directory and they want to move down the directory tree, the user can type the path starting with the name of the next directory down in the directory structure. -

COMP 9 Lab 1: Meeting Kate
COMP 9 Lab 1: Meeting Kate Out: Thursday, January 27th, 1:15 PM Due: Thursday, February 3rd, 1:15 PM 1 Overview This assignment introduces you to the Linux operating system, the ruby in- terpreter, and the Kate text editor. You will use these tools to write a Ruby program and run it. 2 Navigating Linux and Kate (Lab part 1) The computers should already be on; if the screen is blank, simply hit a key on the keyboard and wait a few seconds. You should be able to log in with your recently created CS account. Enter your CS user name and password to log in. Right-click on the desktop and select \Terminal" from the popup menu. A window will appear. This window is running a Linux shell. The shell takes commands typed on the keyboard and executes them. You should see a prompt that looks something like this: lab116s{ndaniels}: This prompt indicates that Linux is ready to execute your next command. You'll use the prompt to launch the Kate text editor, navigate the filesystem, and submit your completed assignments. The filesystem is the hierarchical directory (\folder") system of Linux. Unlike Windows, in which the file hierarchy starts with a drive letter and :\ (for example, C:\), the Linux file system always starts with a /, which is called the root of the file system. When you first start the Terminal, you are in your home directory. Now, create a new directory within your home directory (a subdirectory) to store all of your Comp 9 files for the semester. -

Command Line Commands Cd = Change Directory (.. = Go up a Level)
Command Line Commands Cd = change directory (.. = go up a level) Ls = list of files in current dir Ls -a = list all Ls -l = list files in long format Ls -t = list files in order of last modified Ls -alt = all list options used together Cp = copies files & dir (ex: cp Frida.txt Lincoln.txt -> copies Frida into Lincoln); can also copy multiple files at once (source files first & destination last) Mkdir = make directory Touch = updates “last modified” date to now. Also creates empty file if filename doesn’t exist Mv = move (ex: mv wonderwoman.txt batman.txt superhero/) = moves wonderwoman.txt & batman.txt to the superhero/ folder; can also be used to rename a file Rm = remove; deletes files Rm -r – (“recursive”) deletes a directory and all child directories Pwd = print working directory Exit = close cmd line window *up arrow = repeat last command Echo = command acceps string (ex: $echo “Hello”) as standard input (stdin) and echoes the same string back to the terminal as standard output (stdout). *NOTE: (stderr) is a standard error message for failed process > = (overwrites) command redirects standard output to a file (ex: $echo “Hello” > hello.txt) Cat = commands a file to output the contents to the terminal (ex: $cat hello.txt) >> = (appends) redirects output to file, but appends to content already in file < = takes standard input from file on right & inputs it into program on the left | = (“pipe”) Takes the standard output of command on the left, and pipes it as standard input to the command on the right Sort = takes the standard input and orders it alphabetically for the output Uniq = (“unique”) filters out adjacent, duplicate lines in a file. -
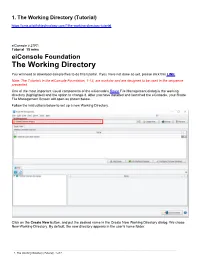
The Working Directory (Tutorial)
1. The Working Directory (Tutorial) https://cms.pilotfishtechnology.com/1the-working-directory-tutorial eiConsole v.21R1 Tutorial 15 mins eiConsole Foundation The Working Directory You will need to download sample files to do this tutorial. If you have not done so yet, please click this LINK. Note: The Tutorials in the eiConsole Foundation, 1-13, are modular and are designed to be used in the sequence presented. One of the most important visual components of the eiConsole’s Route File Management dialog is the working directory (highlighted) and the option to change it. After you have installed and launched the eiConsole, your Route File Management Screen will open as shown below. Follow the instructions below to set up a new Working Directory. Click on the Create New button, and put the desired name in the Create New Working Directory dialog. We chose New-Working-Directory. By default, the new directory appears in the user’s home folder. ______________________________________________________________________________________________________________________________________________________ 1. The Working Directory (Tutorial). 1 of 7 The new empty Working Directory will open. ______________________________________________________________________________________________________________________________________________________ 1. The Working Directory (Tutorial). 2 of 7 To change your working directory click the “Browse” button. The Select Working Directory dialog window opens. It is set to the current New-Working-Directory, and as shown below and the associated filesystem is visible. Use the down arrow to navigate to your default files. Here we see the path to our Getting Started Project files – C:\Program Files\PilotFish Technology\eiConsole\samples\Getting-Started-Project. ______________________________________________________________________________________________________________________________________________________ 1. The Working Directory (Tutorial). 3 of 7 The buttons on the right allow easy navigation as per the Contextual HELP flags. -

Intro to Cluster Computing
Introduction to the Linux Command Line for High-Performance Computing Ken Weiss MSIS Computational Research Consulting Division Roadmap The command shell Navigating the filesystem Basic commands & wildcarding Shell redirection & pipelining Editing text files Permissions Processes Environment variables and customizing your session cja/kgw 2016 2 1/16 Poll How many are here to learn the command line: For HPC, specifically Flux, work? For other computing cluster work? For desktops/other Linux/Unix/Mac OS X? Other? cja 2014 3 10/14 Course Text William E Shotts, Jr., “The Linux Command Line: A Complete Introduction,” No Starch Press, January 2012. Download Creative Commons Licensed version at http://downloads.sourceforge.net/project/linuxcommand /TLCL/13.07/TLCL-13.07.pdf. cja 2014 4 10/14 The command shell cja 2014 5 10/14 What you are used to using kgw 2016 6 1/16 What you will be using kgw 2016 7 1/16 The command shell The command shell is an application that reads command lines from the keyboard and passes them to the Linux operating system to be executed. When you login to a remote Linux system, using a tool like ssh, you will automatically be connected to a shell. Your computing session is kept separate from other user’s computing sessions, because they are “enclosed” in a “shell”. On your desktop, laptop, or tablet, you may have to find and execute a terminal emulator application to bring up a shell in a window. cja 2014 8 10/14 The command shell cja 2014 9 10/14 http://askubuntu.com/questions/161511/are-the-linux-utilities-parts-of-the-kernel-shell