Thermostable Rnase P Rnas Lacking P18 Identified in the Aquificales
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Efficient Genome Editing of an Extreme Thermophile, Thermus
www.nature.com/scientificreports OPEN Efcient genome editing of an extreme thermophile, Thermus thermophilus, using a thermostable Cas9 variant Bjorn Thor Adalsteinsson1*, Thordis Kristjansdottir1,2, William Merre3, Alexandra Helleux4, Julia Dusaucy5, Mathilde Tourigny4, Olafur Fridjonsson1 & Gudmundur Oli Hreggvidsson1,2 Thermophilic organisms are extensively studied in industrial biotechnology, for exploration of the limits of life, and in other contexts. Their optimal growth at high temperatures presents a challenge for the development of genetic tools for their genome editing, since genetic markers and selection substrates are often thermolabile. We sought to develop a thermostable CRISPR-Cas9 based system for genome editing of thermophiles. We identifed CaldoCas9 and designed an associated guide RNA and showed that the pair have targetable nuclease activity in vitro at temperatures up to 65 °C. We performed a detailed characterization of the protospacer adjacent motif specifcity of CaldoCas9, which revealed a preference for 5′-NNNNGNMA. We constructed a plasmid vector for the delivery and use of the CaldoCas9 based genome editing system in the extreme thermophile Thermus thermophilus at 65 °C. Using the vector, we generated gene knock-out mutants of T. thermophilus, targeting genes on the bacterial chromosome and megaplasmid. Mutants were obtained at a frequency of about 90%. We demonstrated that the vector can be cured from mutants for a subsequent round of genome editing. CRISPR-Cas9 based genome editing has not been reported previously in the extreme thermophile T. thermophilus. These results may facilitate development of genome editing tools for other extreme thermophiles and to that end, the vector has been made available via the plasmid repository Addgene. -

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

The Genome of Prasinoderma Coloniale Unveils the Existence of a Third Phylum Within Green Plants
SUPPLEMENTARY INFORMATIONARTICLES https://doi.org/10.1038/s41559-020-1221-7 In the format provided by the authors and unedited. The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants Linzhou Li1,2,13, Sibo Wang1,3,13, Hongli Wang1,4, Sunil Kumar Sahu 1, Birger Marin 5, Haoyuan Li1, Yan Xu1,4, Hongping Liang1,4, Zhen Li 6, Shifeng Cheng1, Tanja Reder5, Zehra Çebi5, Sebastian Wittek5, Morten Petersen3, Barbara Melkonian5,7, Hongli Du8, Huanming Yang1, Jian Wang1, Gane Ka-Shu Wong 1,9, Xun Xu 1,10, Xin Liu 1, Yves Van de Peer 6,11,12 ✉ , Michael Melkonian5,7 ✉ and Huan Liu 1,3 ✉ 1State Key Laboratory of Agricultural Genomics, BGI-Shenzhen, Shenzhen, China. 2Department of Biotechnology and Biomedicine, Technical University of Denmark, Lyngby, Denmark. 3Department of Biology, University of Copenhagen, Copenhagen, Denmark. 4BGI Education Center, University of Chinese Academy of Sciences, Shenzhen, China. 5Institute for Plant Sciences, Department of Biological Sciences, University of Cologne, Cologne, Germany. 6Department of Plant Biotechnology and Bioinformatics (Ghent University) and Center for Plant Systems Biology, Ghent, Belgium. 7Central Collection of Algal Cultures, Faculty of Biology, University of Duisburg-Essen, Essen, Germany. 8School of Biology and Biological Engineering, South China University of Technology, Guangzhou, China. 9Department of Biological Sciences and Department of Medicine, University of Alberta, Edmonton, Alberta, Canada. 10Guangdong Provincial Key Laboratory of Genome Read and Write, BGI-Shenzhen, Shenzhen, China. 11College of Horticulture, Nanjing Agricultural University, Nanjing, China. 12Centre for Microbial Ecology and Genomics, Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria, South Africa. -
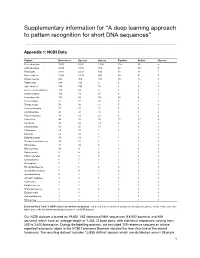
A Deep Learning Approach to Pattern Recognition for Short DNA Sequences”
Supplementary information for “A deep learning approach to pattern recognition for short DNA sequences” Appendix 1: NCBI Data Phylum References Species Genera Families Orders Classes Proteobacteria 7,053 5,061 1,106 158 55 9 Actinobacteria 4,768 3,313 383 68 29 6 Firmicutes 3,814 2,531 499 56 13 7 Bacteroidetes 1,934 1,525 360 39 8 7 Euryarchaeota 834 450 100 28 13 8 Tenericutes 266 195 8 5 4 1 Spirochaetes 146 100 16 6 4 1 Deinococcus-Thermus 118 99 9 3 2 1 Crenarchaeota 113 61 27 8 5 1 Cyanobacteria 113 88 59 30 8 2 Fusobacteria 75 37 10 2 1 1 Thermotogae 70 48 13 5 4 1 Verrucomicrobia 57 53 22 7 4 3 Acidobacteria 45 41 19 5 5 4 Planctomycetes 44 31 23 5 3 2 Chloroflexi 44 35 26 15 12 8 Aquificae 43 32 14 4 2 1 Synergistetes 31 25 15 1 1 1 Chlamydiae 28 18 7 5 2 1 Chlorobi 21 16 5 1 1 1 Deferribacteres 15 11 7 1 1 1 Thermodesulfobacteria 14 12 5 1 1 1 Nitrospirae 11 10 3 1 1 1 Fibrobacteres 10 4 3 3 3 3 Balneolaeota 9 9 4 1 1 1 Chrysiogenetes 6 4 3 1 1 1 Lentisphaerae 6 5 3 3 3 2 Dictyoglomi 5 2 1 1 1 1 Rhodothermaeota 5 5 3 2 1 1 Gemmatimonadetes 5 4 3 2 2 2 Ignavibacteriae 4 2 2 2 1 1 Armatimonadetes 4 3 3 3 3 3 Caldiserica 3 1 1 1 1 1 Calditrichaeota 2 2 1 1 1 1 Thaumarchaeota 2 2 2 2 2 2 Elusimicrobia 2 1 1 1 1 1 Kiritimatiellaeota 1 1 1 1 1 1 Nitrospinae 1 1 1 1 1 1 Extended Data Table 1: NCBI dataset breakdown by phylum. -

7. References
University of Akureyri Department of Natural Resource Science 7. References Ammann, E. C., Reed, L. L., & Durichek, J. J. (1968). Gas consumptions and Growth Rate of Hydrogenomonas eutropha in Continuous Culture. Applied Microbiology , 16, (6), 822-826. Aguiar, P., Beveridge, T. J., & Reysenbach, A.-L. (2004). Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen oxidizing microaerophile from terrestrial hot springs in the Azores. International Journal of Systematic and Evolutionary Microbiology , 54, 33-39. Altschul, S., Gish, W., Miller, W., Myers, E., & Lipman, D. (1990). "Basic local alignment search tool.". J. Mol. Biol. , 215:403-410. Amend, J., & Shock, E. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiology Reviews , 25, 175-243. Aragno, M. (1978). Enrichment, isolation and preliminary characterization of a thermophilic, endospore-forming hydrogen bacterium. FEMS Micobiol. Lett. , 3: 13-15. Aragno, M. (1992). The Thermophilic, Aerobic, Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Trüper, M. Dworkin, W. Harder, & K. Schleifer, The Prokaryotes, a handbook on biology of bacteria. 2nd ed. vol. 4 (pp. 3917-3933.). New York: Springer Verlag. Aragno, M., & Schlegel, H. G. (1992). The mesophilic Hydrogen-Oxidizing (Knallgas) Bacteria. In A. Balows, H. Truper, M. Dworkin, W. Harder, & K.-H. Schleifer, The Prokaryotes 2nd. ed. (pp. 344-384). New York: Springer. Ármannson, H. (2002, May 30-31). Erindi á ráðstefnu um málefni veitufyrirtækja . Grænt bókhald í jarðhita- samanburður á útblæstri við aðra orkugjafa . Akureyri, Iceland: Samorka. Bae, S., Kwak, K., Kim, S., Chung, S., & Igarashi, Y. (2001). Isolation and Characterization of CO2-Fixing Hydrogen -Oxidizing Marine 109 University of Akureyri Department of Natural Resource Science Bacteria. -

Characterization of the Uncommon Enzymes from (2004)
OPEN Mannosylglucosylglycerate biosynthesis SUBJECT AREAS: in the deep-branching phylum WATER MICROBIOLOGY MARINE MICROBIOLOGY Planctomycetes: characterization of the HOMEOSTASIS MULTIENZYME COMPLEXES uncommon enzymes from Rhodopirellula Received baltica 13 March 2013 Sofia Cunha1, Ana Filipa d’Avo´1, Ana Mingote2, Pedro Lamosa3, Milton S. da Costa1,4 & Joana Costa1,4 Accepted 23 July 2013 1Center for Neuroscience and Cell Biology, University of Coimbra, 3004-517 Coimbra, Portugal, 2Instituto de Tecnologia Quı´mica Published e Biolo´gica, Universidade Nova de Lisboa, 2780-157 Oeiras, Portugal, 3Centro de Ressonaˆncia Magne´tica Anto´nio Xavier, 7 August 2013 Instituto de Tecnologia Quı´mica e Biolo´gica, Universidade Nova de Lisboa, 2781-901 Oeiras, Portugal, 4Department of Life Sciences, University of Coimbra, Apartado 3046, 3001-401 Coimbra, Portugal. Correspondence and The biosynthetic pathway for the rare compatible solute mannosylglucosylglycerate (MGG) accumulated by requests for materials Rhodopirellula baltica, a marine member of the phylum Planctomycetes, has been elucidated. Like one of the should be addressed to pathways used in the thermophilic bacterium Petrotoga mobilis, it has genes coding for J.C. ([email protected].) glucosyl-3-phosphoglycerate synthase (GpgS) and mannosylglucosyl-3-phosphoglycerate (MGPG) synthase (MggA). However, unlike Ptg. mobilis, the mesophilic R. baltica uses a novel and very specific MGPG phosphatase (MggB). It also lacks a key enzyme of the alternative pathway in Ptg. mobilis – the mannosylglucosylglycerate synthase (MggS) that catalyses the condensation of glucosylglycerate with GDP-mannose to produce MGG. The R. baltica enzymes GpgS, MggA, and MggB were expressed in E. coli and characterized in terms of kinetic parameters, substrate specificity, temperature and pH dependence. -

And Thermo-Adaptation in Hyperthermophilic Archaea: Identification of Compatible Solutes, Accumulation Profiles, and Biosynthetic Routes in Archaeoglobus Spp
Universidade Nova de Lisboa Osmo- andInstituto thermo de Tecnologia-adaptation Química e Biológica in hyperthermophilic Archaea: Subtitle Subtitle Luís Pedro Gafeira Gonçalves Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. OH OH OH CDP c c c - CMP O O - PPi O3P P CTP O O O OH OH OH OH OH OH O- C C C O P O O P i Dissertation presented to obtain the Ph.D degree in BiochemistryO O- Instituto de Tecnologia Química e Biológica | Universidade Nova de LisboaP OH O O OH OH OH Oeiras, Luís Pedro Gafeira Gonçalves January, 2008 2008 Universidade Nova de Lisboa Instituto de Tecnologia Química e Biológica Osmo- and thermo-adaptation in hyperthermophilic Archaea: identification of compatible solutes, accumulation profiles, and biosynthetic routes in Archaeoglobus spp. This dissertation was presented to obtain a Ph. D. degree in Biochemistry at the Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa. By Luís Pedro Gafeira Gonçalves Supervised by Prof. Dr. Helena Santos Oeiras, January, 2008 Apoio financeiro da Fundação para a Ciência e Tecnologia (POCI 2010 – Formação Avançada para a Ciência – Medida IV.3) e FSE no âmbito do Quadro Comunitário de apoio, Bolsa de Doutoramento com a referência SFRH / BD / 5076 / 2001. ii ACKNOWNLEDGMENTS The work presented in this thesis, would not have been possible without the help, in terms of time and knowledge, of many people, to whom I am extremely grateful. Firstly and mostly, I need to thank my supervisor, Prof. Helena Santos, for her way of thinking science, her knowledge, her rigorous criticism, and her commitment to science. -

Being Aquifex Aeolicus: Untangling a Hyperthermophile's Checkered Past
GBE Being Aquifex aeolicus: Untangling a Hyperthermophile’s Checkered Past Robert J.M. Eveleigh1,2, Conor J. Meehan1,2,JohnM.Archibald1, and Robert G. Beiko2,* 1Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, Nova Scotia, Canada 2Faculty of Computer Science, Dalhousie University, Halifax, Nova Scotia, Canada *Corresponding author: E-mail: [email protected]. Accepted: November 22, 2013 Abstract Lateral gene transfer (LGT) is an important factor contributing to the evolution of prokaryotic genomes. The Aquificae are a hyper- thermophilic bacterial group whose genes show affiliations to many other lineages, including the hyperthermophilic Thermotogae, the Proteobacteria, and the Archaea. Previous phylogenomic analyses focused on Aquifex aeolicus identified Thermotogae and Downloaded from Aquificae either as successive early branches or sisters in a rooted bacterial phylogeny, but many phylogenies and cellular traits have suggested a stronger affiliation with the Epsilonproteobacteria. Different scenarios for the evolution of the Aquificae yield different phylogenetic predictions. Here, we outline these scenarios and consider the fit of the available data, including three sequenced Aquificae genomes, to different sets of predictions. Evidence from phylogenetic profiles and trees suggests that the Epsilonproteobacteria have the strongest affinities with the three Aquificae analyzed. However, this pattern is shown by only a http://gbe.oxfordjournals.org/ minority of encoded proteins, and the Archaea, many lineages of thermophilic bacteria, and members of genus Clostridium and class Deltaproteobacteria also show strong connections to the Aquificae. The phylogenetic affiliations of different functional subsystems showed strong biases: Most but not all genes implicated in the core translational apparatus tended to group Aquificae with Thermotogae, whereas a wide range of metabolic and cellular processes strongly supported the link between Aquificae and Epsilonproteobacteria. -

Counts Metabolic Yr10.Pdf
Advanced Review Physiological, metabolic and biotechnological features of extremely thermophilic microorganisms James A. Counts,1 Benjamin M. Zeldes,1 Laura L. Lee,1 Christopher T. Straub,1 Michael W.W. Adams2 and Robert M. Kelly1* The current upper thermal limit for life as we know it is approximately 120C. Microorganisms that grow optimally at temperatures of 75C and above are usu- ally referred to as ‘extreme thermophiles’ and include both bacteria and archaea. For over a century, there has been great scientific curiosity in the basic tenets that support life in thermal biotopes on earth and potentially on other solar bodies. Extreme thermophiles can be aerobes, anaerobes, autotrophs, hetero- trophs, or chemolithotrophs, and are found in diverse environments including shallow marine fissures, deep sea hydrothermal vents, terrestrial hot springs— basically, anywhere there is hot water. Initial efforts to study extreme thermo- philes faced challenges with their isolation from difficult to access locales, pro- blems with their cultivation in laboratories, and lack of molecular tools. Fortunately, because of their relatively small genomes, many extreme thermo- philes were among the first organisms to be sequenced, thereby opening up the application of systems biology-based methods to probe their unique physiologi- cal, metabolic and biotechnological features. The bacterial genera Caldicellulosir- uptor, Thermotoga and Thermus, and the archaea belonging to the orders Thermococcales and Sulfolobales, are among the most studied extreme thermo- philes to date. The recent emergence of genetic tools for many of these organ- isms provides the opportunity to move beyond basic discovery and manipulation to biotechnologically relevant applications of metabolic engineering. -

The Thermal Limits to Life on Earth
International Journal of Astrobiology 13 (2): 141–154 (2014) doi:10.1017/S1473550413000438 © Cambridge University Press 2014. The online version of this article is published within an Open Access environment subject to the conditions of the Creative Commons Attribution licence http://creativecommons.org/licenses/by/3.0/. The thermal limits to life on Earth Andrew Clarke1,2 1British Antarctic Survey, Cambridge, UK 2School of Environmental Sciences, University of East Anglia, Norwich, UK e-mail: [email protected] Abstract: Living organisms on Earth are characterized by three necessary features: a set of internal instructions encoded in DNA (software), a suite of proteins and associated macromolecules providing a boundary and internal structure (hardware), and a flux of energy. In addition, they replicate themselves through reproduction, a process that renders evolutionary change inevitable in a resource-limited world. Temperature has a profound effect on all of these features, and yet life is sufficiently adaptable to be found almost everywhere water is liquid. The thermal limits to survival are well documented for many types of organisms, but the thermal limits to completion of the life cycle are much more difficult to establish, especially for organisms that inhabit thermally variable environments. Current data suggest that the thermal limits to completion of the life cycle differ between the three major domains of life, bacteria, archaea and eukaryotes. At the very highest temperatures only archaea are found with the current high-temperature limit for growth being 122 °C. Bacteria can grow up to 100 °C, but no eukaryote appears to be able to complete its life cycle above *60 °C and most not above 40 °C. -

Ionizing Radiation Resistance in Deinococcus Radiodurans
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by CSCanada.net: E-Journals (Canadian Academy of Oriental and Occidental Culture,... ISSN 1715-7862 [PRINT] Advances in Natural Science ISSN 1715-7870 [ONLINE] Vol. 7, No. 2, 2014, pp. 6-14 www.cscanada.net DOI: 10.3968/5058 www.cscanada.org Ionizing Radiation Resistance in Deinococcus Radiodurans LI Wei[a]; MA Yun[a]; XIAO Fangzhu[a]; HE Shuya[a],* [a]Institute of Biochemistry and Molecular Biology, University of South Treatment of D. radiodurans with an acute dose of 5,000 China, Hengyang, China. Gy of ionizing radiation with almost no loss of viability, *Corresponding author. and an acute dose of 15,000 Gy with 37% viability (Daly, Supported by the National Natural Science Foundation of China 2009; Ito, Watanabe, Takeshia, & Iizuka, 1983; Moseley (81272993). & Mattingly, 1971). In contrast, 5 Gy of ionizing radiation can kill a human, 200-800 Gy of ionizing radiation will Received 12 March 2014; accepted 2 June 2014 Published online 26 June 2014 kill E. coli, and more than 4,000 Gy of ionizing radiation will kill the radiation-resistant tardigrade. D. radiodurans can survive 5,000 to 30,000 Gy of ionizing radiation, Abstract which breaks its genome into hundreds of fragments (Daly Deinococcus radiodurans is unmatched among all known & Minton, 1995; Minton, 1994; Slade & Radman, 2011). species in its ability to resist ionizing radiation and other Surprisingly, the genome is reassembled accurately before DNA-damaging factors. It is considered a model organism beginning of the next cycle of cell division. -

On the Chimeric Nature, Thermophilic Origin, and Phylogenetic Placement of the Thermotogales
On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales Olga Zhaxybayevaa, Kristen S. Swithersb, Pascal Lapierrec, Gregory P. Fournierb, Derek M. Bickhartb, Robert T. DeBoyd, Karen E. Nelsond, Camilla L. Nesbøe,f, W. Ford Doolittlea,1, J. Peter Gogartenb, and Kenneth M. Nollb aDepartment of Biochemistry and Molecular Biology, Dalhousie University, 5850 College Street, Halifax, Nova Scotia, Canada B3H 1X5; bDepartment of Molecular and Cell Biology, University of Connecticut, Storrs, CT 06269-3125; cBiotechnology-Bioservices Center, University of Connecticut, Storrs, CT 06269-3149; dThe J. Craig Venter Institute, 9704 Medical Center Drive, Rockville, MD 20850; eCentre for Ecological and Evolutionary Synthesis (CEES), Department of Biology, University of Oslo, P.O. Box 1066 Blindern, N-0316 Oslo, Norway; and fDepartment of Biological Sciences, University of Alberta, Edmonton, Alberta, Canada T6G 2E9 Contributed by W. Ford Doolittle, February 11, 2009 (sent for review January 6, 2009) Since publication of the first Thermotogales genome, Thermotoga developments are relevant here: first that mesophilic Thermo- maritima strain MSB8, single- and multi-gene analyses have dis- togales have been discovered (2), raising the possibility that agreed on the phylogenetic position of this order of Bacteria. Here hyperthermophily is not ancestral to the group, and second that we present the genome sequences of 4 additional members of the a thorough analysis of A. aeolicus shows that, although many of Thermotogales (Tt. petrophila, Tt. lettingae, Thermosipho melane- its informational genes support sisterhood with Tt. maritima, siensis, and Fervidobacterium nodosum) and a comprehensive substantial exchange of some of these genes has occurred with comparative analysis including the original T.