Impaired Eif5a Function Causes a Mendelian Disorder That Is Partially Rescued in Model Systems by Spermidine
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Characterization of the Small RNA Transcriptomes of Cell Protrusions and Cell Bodies of Highly Metastatic Hepatocellular Carcinoma Cells Via RNA Sequencing
ONCOLOGY LETTERS 22: 568, 2021 Characterization of the small RNA transcriptomes of cell protrusions and cell bodies of highly metastatic hepatocellular carcinoma cells via RNA sequencing WENPIN CAI1*, JINGZHANG JI2*, BITING WU2*, KAIXUAN HAO2, PING REN2, YU JIN2, LIHONG YANG2, QINGCHAO TONG2 and ZHIFA SHEN2 1Department of Laboratory Medicine, Wen Zhou Traditional Chinese Medicine Hospital; 2Zhejiang Provincial Key Laboratory of Medical Genetics, Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, Zhejiang 325035, P.R. China Received July 11, 2020; Accepted February 23, 2021 DOI: 10.3892/ol.2021.12829 Abstract. Increasing evidence suggest that hepatocellular differentially expressed miRNAs and circRNAs. The interac‑ carcinoma (HCC) HCCLM3 cells initially develop pseudo‑ tion maps between miRNAs and circRNAs were constructed, podia when they metastasize, and microRNAs (miRNAs/miRs) and signaling pathway maps were analyzed to determine the and circular RNAs (circRNAs) have been demonstrated to molecular mechanism and regulation of the differentially serve important roles in the development, progression and expressed miRNAs and circRNAs. Taken together, the results metastasis of cancer. The present study aimed to isolate the of the present study suggest that the Boyden chamber assay cell bodies (CBs) and cell protrusions (CPs) from HCCLM3 can be used to effectively isolate the somatic CBs and CPs of cells, and screen the miRNAs and circRNAs associated with HCC, which can be used to screen the miRNAs and circRNAs HCC infiltration and metastasis in CBs and CPs. The Boyden associated with invasion and metastasis of HCC. chamber assay has been confirmed to effectively isolate the CBs and CPs from HCCLM3 cells via observation of microtu‑ Introduction bule immunofluorescence, DAPI staining and nuclear protein H3 western blotting. -

Relevance of Translation Initiation in Diffuse Glioma Biology and Its
cells Review Relevance of Translation Initiation in Diffuse Glioma Biology and its Therapeutic Potential Digregorio Marina 1, Lombard Arnaud 1,2, Lumapat Paul Noel 1, Scholtes Felix 1,2, Rogister Bernard 1,3 and Coppieters Natacha 1,* 1 Laboratory of Nervous System Disorders and Therapy, GIGA-Neurosciences Research Centre, University of Liège, 4000 Liège, Belgium; [email protected] (D.M.); [email protected] (L.A.); [email protected] (L.P.N.); [email protected] (S.F.); [email protected] (R.B.) 2 Department of Neurosurgery, CHU of Liège, 4000 Liège, Belgium 3 Department of Neurology, CHU of Liège, 4000 Liège, Belgium * Correspondence: [email protected] Received: 18 October 2019; Accepted: 26 November 2019; Published: 29 November 2019 Abstract: Cancer cells are continually exposed to environmental stressors forcing them to adapt their protein production to survive. The translational machinery can be recruited by malignant cells to synthesize proteins required to promote their survival, even in times of high physiological and pathological stress. This phenomenon has been described in several cancers including in gliomas. Abnormal regulation of translation has encouraged the development of new therapeutics targeting the protein synthesis pathway. This approach could be meaningful for glioma given the fact that the median survival following diagnosis of the highest grade of glioma remains short despite current therapy. The identification of new targets for the development of novel therapeutics is therefore needed in order to improve this devastating overall survival rate. This review discusses current literature on translation in gliomas with a focus on the initiation step covering both the cap-dependent and cap-independent modes of initiation. -

Transcriptional Changes Involved in Atrophying Muscles During Prolonged Fasting in Rats
International Journal of Molecular Sciences Article Transcriptional Changes Involved in Atrophying Muscles during Prolonged Fasting in Rats 1,2 1,2, 3 1,2 Marianne Ibrahim , Thierry Wasselin y, Etienne Challet , Alain Van Dorsselaer , Yvon Le Maho 1,4,5, Thierry Raclot 1,4 and Fabrice Bertile 1,2,* 1 Institut Pluridisciplinaire Hubert Curien (IPHC), CNRS, Université de Strasbourg, 67000 Strasbourg, France; [email protected] (M.I.); [email protected] (T.W.); [email protected] (A.V.D.); [email protected] (Y.L.M.); [email protected] (T.R.) 2 Laboratoire de Spectrométrie de Masse Bio-Organique, 25 rue Becquerel, F-67087 Strasbourg, France 3 Institute of Cellular and Integrative Neurosciences, CNRS, Université de Strasbourg, F-67000 Strasbourg, France; [email protected] 4 Département Ecologie, Physiologie, Ethologie, 23 rue Becquerel, F-67087 Strasbourg, France 5 Centre Scientifique de Monaco, 8 quai Antoine 1er, 98000 Monaco, Monaco * Correspondence: [email protected]; Tel.: +33-3-68-85-26-81 Present address: Department of Clinical Chemistry, University Medical Center, 37075 Göttingen, Germany. y Received: 3 July 2020; Accepted: 18 August 2020; Published: 20 August 2020 Abstract: Food deprivation resulting in muscle atrophy may be detrimental to health. To better understand how muscle mass is regulated during such a nutritional challenge, the current study deciphered muscle responses during phase 2 (P2, protein sparing) and phase 3 (P3, protein mobilization) of prolonged fasting in rats. This was done using transcriptomics analysis and a series of biochemistry measurements. The main findings highlight changes for plasma catabolic and anabolic stimuli, as well as for muscle transcriptome, energy metabolism, and oxidative stress. -
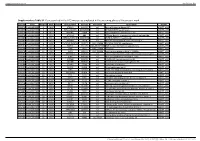
Supplementary Table S1. Genes Printed in the HC5 Microarray Employed in the Screening Phase of the Present Work
Supplementary material Ann Rheum Dis Supplementary Table S1. Genes printed in the HC5 microarray employed in the screening phase of the present work. CloneID Plate Position Well Length GeneSymbol GeneID Accession Description Vector 692672 HsxXG013989 2 B01 STK32A 202374 null serine/threonine kinase 32A pANT7_cGST 692675 HsxXG013989 3 C01 RPS10-NUDT3 100529239 null RPS10-NUDT3 readthrough pANT7_cGST 692678 HsxXG013989 4 D01 SPATA6L 55064 null spermatogenesis associated 6-like pANT7_cGST 692679 HsxXG013989 5 E01 ATP1A4 480 null ATPase, Na+/K+ transporting, alpha 4 polypeptide pANT7_cGST 692689 HsxXG013989 6 F01 ZNF816-ZNF321P 100529240 null ZNF816-ZNF321P readthrough pANT7_cGST 692691 HsxXG013989 7 G01 NKAIN1 79570 null Na+/K+ transporting ATPase interacting 1 pANT7_cGST 693155 HsxXG013989 8 H01 TNFSF12-TNFSF13 407977 NM_172089 TNFSF12-TNFSF13 readthrough pANT7_cGST 693161 HsxXG013989 9 A02 RAB12 201475 NM_001025300 RAB12, member RAS oncogene family pANT7_cGST 693169 HsxXG013989 10 B02 SYN1 6853 NM_133499 synapsin I pANT7_cGST 693176 HsxXG013989 11 C02 GJD3 125111 NM_152219 gap junction protein, delta 3, 31.9kDa pANT7_cGST 693181 HsxXG013989 12 D02 CHCHD10 400916 null coiled-coil-helix-coiled-coil-helix domain containing 10 pANT7_cGST 693184 HsxXG013989 13 E02 IDNK 414328 null idnK, gluconokinase homolog (E. coli) pANT7_cGST 693187 HsxXG013989 14 F02 LYPD6B 130576 null LY6/PLAUR domain containing 6B pANT7_cGST 693189 HsxXG013989 15 G02 C8orf86 389649 null chromosome 8 open reading frame 86 pANT7_cGST 693194 HsxXG013989 16 H02 CENPQ 55166 -

Downregulation of Carnitine Acyl-Carnitine Translocase by Mirnas
Page 1 of 288 Diabetes 1 Downregulation of Carnitine acyl-carnitine translocase by miRNAs 132 and 212 amplifies glucose-stimulated insulin secretion Mufaddal S. Soni1, Mary E. Rabaglia1, Sushant Bhatnagar1, Jin Shang2, Olga Ilkayeva3, Randall Mynatt4, Yun-Ping Zhou2, Eric E. Schadt6, Nancy A.Thornberry2, Deborah M. Muoio5, Mark P. Keller1 and Alan D. Attie1 From the 1Department of Biochemistry, University of Wisconsin, Madison, Wisconsin; 2Department of Metabolic Disorders-Diabetes, Merck Research Laboratories, Rahway, New Jersey; 3Sarah W. Stedman Nutrition and Metabolism Center, Duke Institute of Molecular Physiology, 5Departments of Medicine and Pharmacology and Cancer Biology, Durham, North Carolina. 4Pennington Biomedical Research Center, Louisiana State University system, Baton Rouge, Louisiana; 6Institute for Genomics and Multiscale Biology, Mount Sinai School of Medicine, New York, New York. Corresponding author Alan D. Attie, 543A Biochemistry Addition, 433 Babcock Drive, Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin, (608) 262-1372 (Ph), (608) 263-9608 (fax), [email protected]. Running Title: Fatty acyl-carnitines enhance insulin secretion Abstract word count: 163 Main text Word count: 3960 Number of tables: 0 Number of figures: 5 Diabetes Publish Ahead of Print, published online June 26, 2014 Diabetes Page 2 of 288 2 ABSTRACT We previously demonstrated that micro-RNAs 132 and 212 are differentially upregulated in response to obesity in two mouse strains that differ in their susceptibility to obesity-induced diabetes. Here we show the overexpression of micro-RNAs 132 and 212 enhances insulin secretion (IS) in response to glucose and other secretagogues including non-fuel stimuli. We determined that carnitine acyl-carnitine translocase (CACT, Slc25a20) is a direct target of these miRNAs. -

The Neuroprotective Role of the GM1 Oligosaccharide, Ii3neu5ac-Gg4, In
Molecular Neurobiology (2019) 56:6673–6702 https://doi.org/10.1007/s12035-019-1556-8 The Neuroprotective Role of the GM1 Oligosaccharide, 3 II Neu5Ac-Gg4, in Neuroblastoma Cells Elena Chiricozzi1 & Margherita Maggioni1 & Erika di Biase1 & Giulia Lunghi1 & Maria Fazzari1 & Nicoletta Loberto 1 & Maffioli Elisa2 & Francesca Grassi Scalvini2 & Gabriella Tedeschi 2,3 & Sandro Sonnino1 Received: 10 January 2019 /Accepted: 13 March 2019 /Published online: 26 March 2019 # Springer Science+Business Media, LLC, part of Springer Nature 2019 Abstract 3 Recently, we demonstrated that the GM1 oligosaccharide, II Neu5Ac-Gg4 (OligoGM1), administered to cultured murine Neuro2a neuroblastoma cells interacts with the NGF receptor TrkA, leading to the activation of the ERK1/2 downstream pathway and to cell differentiation. To understand how the activation of the TrkA pathway is able to trigger key biochemical signaling, we performed a proteomic analysis on Neuro2a cells treated with 50 μM OligoGM1 for 24 h. Over 3000 proteins were identified. Among these, 324 proteins were exclusively expressed in OligoGM1-treated cells. Interestingly, several proteins expressed only in OligoGM1-treated cells are involved in biochemical mechanisms with a neuroprotective potential, reflecting the GM1 neuroprotective effect. In addition, we found that the exogenous administration of OligoGM1 reduced the cellular oxidative stress in Neuro2a cells and conferred protection against MPTP neurotoxicity. These results confirm and reinforce the idea that the molecular mechanisms underlying the GM1 neurotrophic and neuroprotective effects depend on its oligosaccharide chain, suggesting the activation of a positive signaling starting at plasma membrane level. Keywords GM1 ganglioside . GM1 oligosaccharide chain . TrkA neurotrophin receptor . Plasma membrane signaling . Neuroprotection . -

Expression of EIF2 Family Genes Under Newcastle Disease Virus Infection in Two Chicken Lines
Iowa State University Animal Industry Report 2019 Expression of EIF2 Family Genes under Newcastle Disease Virus Infection in Two Chicken Lines A.S. Leaflet R3323 controlling immune response to NDV. We hypothesize that the lines differentially regulate the expression of genes Ana Paula Del Vesco, Visiting Scholar1,2; related to host protein synthesis as a defense mechanism in Melissa S. Monson, Postdoc Research Associate1; response to NDV infection, and that this genetic control Michael G. Kaiser, Research Associate1; may contribute to the level of host resistance to NDV. Huaijun Zhou, Professor3; Jack C. M. Dekkers, Distinguished Professor1; Materials and Methods Susan J. Lamont, Distinguished Professor1 The effect of NDV challenge on gene expression in the 1Iowa State University, USA; 2Universidade Federal de spleen of the Fayoumis and the Leghorns at two- and six- Sergipe, Brazil; 3University of California, Davis, USA. days post infection (dpi) was evaluated. Chickens from both lines were either challenged with a concentrated NDV Summary and Implications vaccine strain via the eyes and nostrils or non-challenged Newcastle Disease Virus (NDV) causes large economic (control). The chickens from both lines and challenge losses around the world and recent outbreaks have occurred statuses (NDV challenged or control) were euthanized at 2- in the US. We assessed the expression of eukaryotic or 6-dpi for spleen harvest. The RNA was isolated from translation initiation factor 2 (eIF2)-related genes in the spleen samples and used to assess the expression of seven spleens of two genetically different, inbred chicken lines genes related to the eIF2 family using quantitative PCR. The from the Iowa State University Poultry Genetics program. -

Supplemental Material For
SUPPLEMENTAL MATERIAL FOR Coexpression network based on natural variation in human gene expression reveals gene interactions and functions Renuka Nayak, Michael Kearns, Richard S. Spielman, Vivian G. Cheung Supplementary Figure 1 Supplementary Table 1. Gene pairs whose correlations in gene expression levels differ significantly (Pc<0.05) among the 3 datasets. Supplementary Table 2. Gene pairs that are correlated in gene expression levels with |R|>0.5 and are found within 500 kb of each other. Supplementary Table 3. Predicted functions of poorly characterized genes based on the functions of neighboring genes. Supplementary Figure 1. Genes identified in genome-wide association studies (grey) and their neighbors in the network. Red and green connections refer to positive and negative correlations, respectively. MICB has been implicated in AIDS progression (PMID: 19115949) TNF has been implicated in AIDS progression (PMID: 19115949) LTB has been implicated in AIDS progression (PMID: 19115949) ZNF224 has been implicated in Alzheimer's disease (PMID: 19118814) NDUFAB1 has been implicated in bipolar disorder (PMID: 17554300) SFRS10 has been implicated in body mass index (PMID: 19079260) and weight (PMID: 19079260) CTNNBL1 has been implicated in bone mineral density (PMID: 17903296) TGFBR3 has been implicated in bone mineral density (PMID: 19249006) IGF2R has been implicated in brain lesion load (PMID: 19010793) LSP1 has been implicated in breast cancer (PMID: 17529967) FBN1 has been implicated in breast cancer (PMID: 17903305) GLG1 has been implicated in breast cancer (PMID: 18463975) SCHIP1 has been implicated in Celiac disease (PMID: 18311140) RGS1 has been implicated in Celiac disease (PMID: 18311140) FADS2 has been implicated in Cholesterol (total) (PMID: 19060911), HDL cholesterol (PMID: 19060911, 19060906), LDL cholesterol (PMID: 19060911, 19060910), and triglycerides (PMID: 19060906). -

Supplementary Figure 1 Standardization of Gene Expression
Supplementary Figure 1 Standardization of gene expression Notes: (A) Standardization of GSE86544, (B) standardization of GSE103479, (C) standardization of GSE102238, (D) Standardization of GSE7055. The blue bar represents the data before normalization, and the red bar represents the data after normalization. Supplementary Figure 2 Correlation between module eigengenes and clinical traits especially PNI in GSE103479 and GSE102238 datasets. Notes: (A, B) Module-trait relationships in GSE103479 and GSE102238 datasets. The correlation coefficients and corresponding P-values in the brackets are contained in each cell. The table is color- coded by correlation between eigengenes and traits according to the color legend on the right side. The modules with the most significant differences are displayed in brackets. Abbreviations: PNI, perineural invasion. Supplementary Figure 3 The expression values of CCNB2 in pancreatic cancer (GSE102238) and colon cancer (GSE103479). Notes: (A, B) CCNB2 expression values were detected in GSE102238 and GSE103479. Abbreviations: CCNB2, cyclin B2 Supplementary Table 1 Results of top 20 pathway enrichment analysis of GSE7055 Term Category Description Count Log10(P) Genes GO:0000280 GO Biological nuclear division 33 -23.4 BIRC5,BUB1B,CCNB1,CCNE1,CDC20, Processes CKS2,KIF11,MAD2L1,MYBL2,SPAST, TOP2A,TTK,PRC1,PKMYT1,PTTG1,T RIP13,DLGAP5,TACC3,SMC2,SPAG5, UBE2C,ZWINT,TPX2,FBXO5,RACGA P1,NUSAP1,SPDL1,CDCA8,CEP55,ND C1,NSFL1C,KIF18B,ASPM GO:1902850 GO Biological microtubule 15 -12.89 BIRC5,CCNB1,CDC20,KIF11,MAD2L1 Processes -

Eukaryotic Translation Initiation Factors As Promising Targets in Cancer Therapy
Hao et al. Cell Communication and Signaling (2020) 18:175 https://doi.org/10.1186/s12964-020-00607-9 REVIEW Open Access Eukaryotic translation initiation factors as promising targets in cancer therapy Peiqi Hao1,2†, Jiaojiao Yu1†, Richard Ward3, Yin Liu2, Qiao Hao2,SuAn2* and Tianrui Xu2* Abstract The regulation of the translation of messenger RNA (mRNA) in eukaryotic cells is critical for gene expression, and occurs principally at the initiation phase which is mainly regulated by eukaryotic initiation factors (eIFs). eIFs are fundamental for the translation of mRNA and as such act as the primary targets of several signaling pathways to regulate gene expression. Mis-regulated mRNA expression is a common feature of tumorigenesis and the abnormal activity of eIF complexes triggered by upstream signaling pathways is detected in many tumors, leading to the selective translation of mRNA encoding proteins involved in tumorigenesis, metastasis, or resistance to anti-cancer drugs, and making eIFs a promising therapeutic target for various types of cancers. Here, we briefly outline our current understanding of the biology of eIFs, mainly focusing on the effects of several signaling pathways upon their functions and discuss their contributions to the initiation and progression of tumor growth. An overview of the progress in developing agents targeting the components of translation machinery for cancer treatment is also provided. Keywords: eIF, mRNA translation, Cancer, MAPK, PI3K/Akt, mTOR Background eukaryotes utilize many more initiation factors than do pro- The regulation of gene expression in eukaryotes can occur karyotes, reflecting the greater biological complexity of at different stages including gene transcription and mRNA eukaryotic translation. -

Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared Amongst Δγ T, Innate Lymphoid, and Th Cells
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 δγ is online at: average * The Journal of Immunology , 10 of which you can access for free at: 2016; 197:1460-1470; Prepublished online 6 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600643 http://www.jimmunol.org/content/197/4/1460 Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst T, Innate Lymphoid, and Th Cells You Jeong Lee, Gabriel J. Starrett, Seungeun Thera Lee, Rendong Yang, Christine M. Henzler, Stephen C. Jameson and Kristin A. Hogquist J Immunol cites 41 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2016/07/06/jimmunol.160064 3.DCSupplemental This article http://www.jimmunol.org/content/197/4/1460.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 26, 2021. The Journal of Immunology Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst gd T, Innate Lymphoid, and Th Cells You Jeong Lee,* Gabriel J.