Manipulating Fibroblast Environment to Study Specific Gene Expression Elizabeth Mosqueda Boise State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Anti-ITGAE (GW21349)
3050 Spruce Street, Saint Louis, MO 63103 USA Tel: (800) 521-8956 (314) 771-5765 Fax: (800) 325-5052 (314) 771-5757 email: [email protected] Product Information Anti-ITGAE antibody produced in chicken, affinity isolated antibody Catalog Number GW21349 Formerly listed as GenWay Catalog Number 15-288-21349, Integrin alpha-E Antibody. – Storage Temperature Store at 20 °C The product is a clear, colorless solution in phosphate buffered saline, pH 7.2, containing 0.02% sodium azide. Synonyms: Integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide), Mucosal Species Reactivity: Human lymphocyte 1 antigen; HML-1 antigen; Integrin alpha-IEL; Tested Applications: WB CD103 antigen Recommended Dilutions: Recommended starting dilution Product Description for Western blot analysis is 1:500, for tissue or cell staining Integrin alpha-E/beta-7 is a receptor for E-cadherin. It 1:200. mediates adhesion of intra-epithelial T-lymphocytes to epithelial cell monolayers. Note: Optimal concentrations and conditions for each application should be determined by the user. NCBI Accession number: NP_002199.2 Swiss Prot Accession number: P38570 Precautions and Disclaimer This product is for R&D use only, not for drug, household, or Gene Information: Human .. ITGAE (3682) other uses. Due to the sodium azide content a material Immunogen: Recombinant protein Integrin, alpha E (anti- safety data sheet (MSDS) for this product has been sent to gen CD103, human mucosal lymphocyte antigen 1; alpha the attention of the safety officer of your institution. Please polypeptide) consult the Material Safety Data Sheet for information regarding hazards and safe handling practices. Immunogen Sequence: GI # 6007851, sequence 773 - 817 Storage/Stability For continuous use, store at 2–8 °C for up to one week. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Human CD Marker Chart Reviewed by HLDA1 Bdbiosciences.Com/Cdmarkers
BD Biosciences Human CD Marker Chart Reviewed by HLDA1 bdbiosciences.com/cdmarkers 23-12399-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD93 C1QR1,C1qRP, MXRA4, C1qR(P), Dj737e23.1, GR11 – – – – – + + – – + – CD220 Insulin receptor (INSR), IR Insulin, IGF-2 + + + + + + + + + Insulin-like growth factor 1 receptor (IGF1R), IGF-1R, type I IGF receptor (IGF-IR), CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD94 KLRD1, Kp43 HLA class I, NKG2-A, p39 + – + – – – – – – CD221 Insulin-like growth factor 1 (IGF-I), IGF-II, Insulin JTK13 + + + + + + + + + CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD178, FASLG, APO-1, FAS, TNFRSF6, CD95L, APT1LG1, APT1, FAS1, FASTM, CD95 CD178 (Fas ligand) + + + + + – – IGF-II, TGF-b latency-associated peptide (LAP), Proliferin, Prorenin, Plasminogen, ALPS1A, TNFSF6, FASL Cation-independent mannose-6-phosphate receptor (M6P-R, CIM6PR, CIMPR, CI- CD1d R3G1, R3 b-2-Microglobulin, MHC II CD222 Leukemia -

Role of ITGAE in the Development of Autoimmune Diabetes in Non-Obese Diabetic Mice
E S BARRIE and others Role of ITGAE in autoimmune 224:3 235–243 Research diabetes Role of ITGAE in the development of autoimmune diabetes in non-obese diabetic mice Elizabeth S Barrie, Mels Lodder, Paul H Weinreb1, Jill Buss, Amer Rajab, Christopher Adin2, Qing-Sheng Mi3 and Gregg A Hadley The Ohio State University Wexner Medical Center, Room 216 Tzagournis Medical Research Facility, Correspondence 420 W 12th Avenue, Columbus, Ohio 43201, USA should be addressed 1Biogen Idec, Cambridge, Massachusetts 02142, USA to G A Hadley 2College of Veterinary Medicine, Columbus, Ohio 43201 USA Email 3Henry Ford Hospital, Detroit, Michigan 48202, USA [email protected] Abstract C There is compelling evidence that autoreactive CD8 Tcells play a central role in precipitating Key Words the development of autoimmune diabetes in non-obese diabetic (NOD) mice, but the " diabetes underlying mechanisms remain unclear. Given that ITGAE (CD103) recognizes an islet- " autoimmune restricted ligand (E-cadherin), we postulated that its expression is required for initiation of " immune system disease. We herein use a mouse model of autoimmune diabetes (NOD/ShiLt mice) to test this " mouse C hypothesis. We demonstrate that ITGAE is expressed by a discrete subset of CD8 T cells that infiltrate pancreatic islets before the development of diabetes. Moreover, we demonstrate that development of diabetes in Itgae-deficient NOD mice is significantly delayed at early Journal of Endocrinology but not late time points, indicating that ITGAE is preferentially involved in early diabetes development. To rule out a potential contribution by closely linked loci to this delay, we treated WT NOD mice beginning at 2 weeks of age through 5 weeks of age with a depleting anti-ITGAE mAb and found a decreased incidence of diabetes following anti-ITGAE mAb treatment compared with mice that received isotype control mAbs or non-depleting mAbs to ITGAE. -
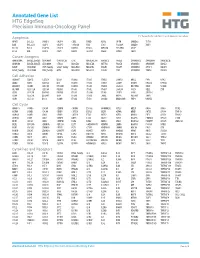
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

ITGAE Rabbit Pab
Leader in Biomolecular Solutions for Life Science ITGAE Rabbit pAb Catalog No.: A9934 Basic Information Background Catalog No. Integrins are heterodimeric integral membrane proteins composed of an alpha chain A9934 and a beta chain. This gene encodes an I-domain-containing alpha integrin that undergoes post-translational cleavage in the extracellular domain, yielding disulfide- Observed MW linked heavy and light chains. In combination with the beta 7 integrin, this protein forms 110kDa the E-cadherin binding integrin known as the human mucosal lymphocyte-1 antigen. This protein is preferentially expressed in human intestinal intraepithelial lymphocytes Calculated MW (IEL), and in addition to a role in adhesion, it may serve as an accessory molecule for IEL 130kDa activation. Category Primary antibody Applications WB,IHC Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 3682 P38570 IHC 1:50 - 1:200 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 19-200 of human ITGAE (NP_002199.3). Synonyms ITGAE;CD103;HUMINAE Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using ITGAE antibody (A9934) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. Detection: ECL Basic Kit (RM00020). Exposure time: 60s. Antibody | Protein | ELISA Kits | Enzyme | NGS | Service For research use only. -

CD103)-Deficient Mice
Mucosal T Lymphocyte Numbers Are Selectively Reduced in Integrin αE (CD103)-Deficient Mice This information is current as Michael P. Schön, Anu Arya, Elizabeth A. Murphy, of September 24, 2021. Cassandra M. Adams, Ulrike G. Strauch, William W. Agace, Jan Marsal, John P. Donohue, Helen Her, David R. Beier, Sara Olson, Leo Lefrancois, Michael B. Brenner, Michael J. Grusby and Christina M. Parker J Immunol 1999; 162:6641-6649; ; Downloaded from http://www.jimmunol.org/content/162/11/6641 References This article cites 43 articles, 17 of which you can access for free at: http://www.jimmunol.org/content/162/11/6641.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 24, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 1999 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Mucosal T Lymphocyte Numbers Are Selectively Reduced in a 1 Integrin E (CD103)-Deficient Mice Michael P. Scho¨n,2* Anu Arya,* Elizabeth A. -

CD103 / ITGAE Antibody
FOR RESEARCH USE ONLY! 02/21 CD103 / ITGAE Antibody CATALOG NO.: A2300-50 (50 µg) A2300-100 (100 µg) BACKGROUND DESCRIPTION: ITGAE, also known as CD103, is an integrin that pairs with the beta 7 integrin to form the human mucosal lymphocyte-1 antigen. ITGAE is preferentially expressed in human intestinal intraepithelial lymphocytes (IEL). It has a role in adhesion and may serve as an accessory molecule for IEL activation. ITGAE has been associated with diseases such as hairy cell leukemia and splenic marginal zone lymphoma. The presence of cytotoxic T cells positive for CD103 expression has been linked to prolonged survival in several malignancies, including gastric cancer, and is currently being explored as a prognostic biomarker as well as a potential therapeutic target. ITGAE; Mucosal Lymphocyte 1 Antigen; HML-1 Antigen; Integrin Subunit Alpha E; HUMINAE ALTERNATE NAMES: ANTIBODY TYPE: Rabbit polyclonal HOST/ISOTYPE: Rabbit / IgG IMMUNOGEN: Human CD103 peptide sequence (amino acids 19 to 200) PURIFICATION: Affinity purification MOLECULAR WEIGHT: 130 kDa calculated; 110 kDa observed FORM: Liquid FORMULATION: In PBS with 0.02% sodium azide, 50% glycerol, pH 7.3 SPECIES REACTIVITY: Human, Mouse, Rat STORAGE CONDITIONS: Store at -20 °C. Avoid repeated freeze-thaw cycles APPLICATIONS: Western Blot (WB): 1:500 to 1:2000 dilution This information is only intended as a guide. The optimal dilutions must be determined by the user Western blot analysis of lysates prepared from various cell lines using CD103 antibody RELATED PRODUCTS: Anti-α4β7 Integrin (Vedolizumab), Humanized Antibody (Cat. No. A2140) Anti-Alpha 5 Beta 1 Integrin (Volociximab), Human IgG4 Antibody (Cat. -

Neural Cadherin Plays Distinct Roles for Neuronal Survival and Axon Growth Under Different Regenerative Conditions
Research Article: New Research | Disorders of the Nervous System Neural Cadherin Plays Distinct Roles for Neuronal Survival and Axon Growth under Different Regenerative Conditions https://doi.org/10.1523/ENEURO.0325-20.2020 Cite as: eNeuro 2020; 10.1523/ENEURO.0325-20.2020 Received: 21 July 2020 Revised: 26 August 2020 Accepted: 28 August 2020 This Early Release article has been peer-reviewed and accepted, but has not been through the composition and copyediting processes. The final version may differ slightly in style or formatting and will contain links to any extended data. Alerts: Sign up at www.eneuro.org/alerts to receive customized email alerts when the fully formatted version of this article is published. Copyright © 2020 Ribeiro et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license, which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed. 1 Neural Cadherin Plays Distinct Roles for Neuronal Survival and Axon Growth under 2 Different Regenerative Conditions 3 Abbreviated Title: NCAD’s role in neuronal survival and regeneration. 4 Authors: Márcio Ribeiro, Konstantin Levay1, Benito Yon1, Ana Ayupe1, Yadira Salgueiro1, Kevin 5 K. Park1 6 Affiliation: 1Miami Project to Cure Paralysis, Department of Neurosurgery, University of Miami 7 Miller School of Medicine, 1095 NW 14th Ter. Miami, FL 33136 8 Corresponding Author: Kevin K. Park, [email protected] 9 Number of Pages: 38 10 Number of Figures: 9 11 Number of Words for Abstract: 250 12 Number of Words for Introduction: 645 13 Number of Words for Discussion: 2,273 14 Conflict of Interest: The authors declare no competing financial interests. -

Departments of 3Oncology and 4Pathology, St. Jude
UNIVERSAL MONITORING OF MINIMAL RESIDUAL DISEASE IN ACUTE MYELOID LEUKEMIA Elaine Coustan-Smith,1 Guangchun Song,4 Sheila Shurtleff,4 Allen Eng-Juh Yeoh,1,2 Chng Wee Joo,2 Siew Peng Chen,1 Jeffrey E. Rubnitz,3,5 Ching-Hon Pui,3,4,5 James R. Downing,4,5 Dario Campana1,2 1Department of Pediatrics and 2National University Cancer Institute Singapore, National University of Singapore, Singapore; Departments of 3Oncology and 4Pathology, St. Jude Children's Research Hospital, Memphis, TN; and 5University of Tennessee Health Science Center, Memphis, TN Supplemental Material (Supplemental Tables S3-S6; Supplemental Figures S1-S6) 1 Table S3. Genes overexpressed in AML “stem cells” according to previous studies and their overexpression in AML according to the present analysis Gene overexpressed in AML Gene overexpressed in AML AML cases with stem cells according to Saito et stem cells according to Kikushige overexpression in this al.(1) et al.(2) study (%)a WT1 84.7 CD32 CD32 76.4 DOK2 70.1 CD96 66.9 HCK 65.6 CD86 CD86 64.3 CD44 64.3 CD93 56.7 ITGB2, CD18 ITGB2, CD18 52.9 CSF1R, CD115 51.0 IL2RA, CD25 IL2RA, CD25 47.8 LY86 43.3 IL7R, CD127 42.0 CD99 42.0 IL17R 39.5 CD97 CD97 37.6 CD33 CD33 36.9 CD9 36.9 CD1C 35.7 AK5 33.1 BIK 31.2 CD47 30.6 TNFRSF4, CD134 29.3 CD84 29.3 IL2RG, CD132 27.4 ITGB7 27.4 CEACAM6, CD66c 26.8 FLT3 25.5 CD180 <25 CTSC <25 PDE9A <25 CD24 <25 CD36 <25 CD123 <25 ITGAE <25 2 LRG1 Not on HG-U133A array SUCNR1 Not on HG-U133A array TNFSF13B, CD257 Not on HG-U133A array CD366, HAVCR2, TIM-3 Not on HG-U133A array CD371, CLEC12A, CLL-1 Not on HG-U133A array aGene expression was studied by HG-U133A oligonucleotide microarrays in157 AML samples and 7 samples of normal CD34+ myeloid progenitors. -

Tumour Heterogeneity and Intercellular Networks of Nasopharyngeal Carcinoma at Single Cell Resolution
ARTICLE https://doi.org/10.1038/s41467-021-21043-4 OPEN Tumour heterogeneity and intercellular networks of nasopharyngeal carcinoma at single cell resolution Yang Liu1,11, Shuai He 1,2,3,11, Xi-Liang Wang4,11, Wan Peng1, Qiu-Yan Chen1, Dong-Mei Chi1,5, Jie-Rong Chen1,6, Bo-Wei Han1, Guo-Wang Lin1,7, Yi-Qi Li1, Qian-Yu Wang 1, Rou-Jun Peng1, Pan-Pan Wei1, Xiang Guo1, ✉ ✉ Bo Li 8,9, Xiaojun Xia 1, Hai-Qiang Mai1, Xue-Da Hu4, Zemin Zhang 4,10 , Yi-Xin Zeng 1 & ✉ Jin-Xin Bei 1,2,3 1234567890():,; The heterogeneous nature of tumour microenvironment (TME) underlying diverse treatment responses remains unclear in nasopharyngeal carcinoma (NPC). Here, we profile 176,447 cells from 10 NPC tumour-blood pairs, using single-cell transcriptome coupled with T cell receptor sequencing. Our analyses reveal 53 cell subtypes, including tumour-infiltrating CD8+ T, regulatory T (Treg), and dendritic cells (DCs), as well as malignant cells with different Epstein-Barr virus infection status. Trajectory analyses reveal exhausted CD8+ T and immune-suppressive TNFRSF4+ Treg cells in tumours might derive from peripheral CX3CR1+CD8+ T and naïve Treg cells, respectively. Moreover, we identify immune- regulatory and tolerogenic LAMP3+ DCs. Noteworthily, we observe intensive inter-cell interactions among LAMP3+ DCs, Treg, exhausted CD8+ T, and malignant cells, suggesting potential cross-talks to foster an immune-suppressive niche for the TME. Collectively, our study uncovers the heterogeneity and interacting molecules of the TME in NPC at single-cell resolution, which provide insights into the mechanisms underlying NPC progression and the development of precise therapies for NPC.