Modeling Uric Acid Kidney Stones Disease in D. Melanogaster Using Rnai and Dietary Modulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Juvenile Spondyloarthropathies: Inflammation in Disguise
PP.qxd:06/15-2 Ped Perspectives 7/25/08 10:49 AM Page 2 APEDIATRIC Volume 17, Number 2 2008 Juvenile Spondyloarthropathieserspective Inflammation in DisguiseP by Evren Akin, M.D. The spondyloarthropathies are a group of inflammatory conditions that involve the spine (sacroiliitis and spondylitis), joints (asymmetric peripheral Case Study arthropathy) and tendons (enthesopathy). The clinical subsets of spondyloarthropathies constitute a wide spectrum, including: • Ankylosing spondylitis What does spondyloarthropathy • Psoriatic arthritis look like in a child? • Reactive arthritis • Inflammatory bowel disease associated with arthritis A 12-year-old boy is actively involved in sports. • Undifferentiated sacroiliitis When his right toe starts to hurt, overuse injury is Depending on the subtype, extra-articular manifestations might involve the eyes, thought to be the cause. The right toe eventually skin, lungs, gastrointestinal tract and heart. The most commonly accepted swells up, and he is referred to a rheumatologist to classification criteria for spondyloarthropathies are from the European evaluate for possible gout. Over the next few Spondyloarthropathy Study Group (ESSG). See Table 1. weeks, his right knee begins hurting as well. At the rheumatologist’s office, arthritis of the right second The juvenile spondyloarthropathies — which are the focus of this article — toe and the right knee is noted. Family history is might be defined as any spondyloarthropathy subtype that is diagnosed before remarkable for back stiffness in the father, which is age 17. It should be noted, however, that adult and juvenile spondyloar- reported as “due to sports participation.” thropathies exist on a continuum. In other words, many children diagnosed with a type of juvenile spondyloarthropathy will eventually fulfill criteria for Antinuclear antibody (ANA) and rheumatoid factor adult spondyloarthropathy. -
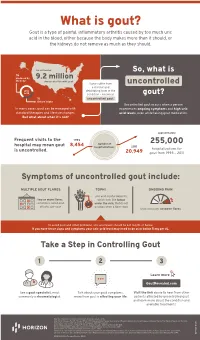
Uncontrolled Gout Fact Sheet
What is gout? Gout is a type of painful, inflammatory arthritis caused by too much uric acid in the blood, either because the body makes more than it should, or the kidneys do not remove as much as they should. An estimated So, what is ⅔ produced by 9.2 million the body Americans live with gout Some suffer from uncontrolled a chronic and uric debilitating form of the acid condition – known as gout? ⅓ uncontrolled gout. dietary intake Uncontrolled gout occurs when a person In many cases, gout can be managed with experiences ongoing symptoms and high uric standard therapies and lifestyle changes. acid levels, even while taking gout medication. But what about when it’s not? approximately Frequent visits to the 1993 number of 255,000 hospital may mean gout 8,454 hospitalizations 2011 is uncontrolled. hospitalizations for 20,949 gout from 1993 – 2011 Symptoms of uncontrolled gout include: MULTIPLE GOUT FLARES TOPHI ONGOING PAIN uric acid crystal deposits, two or more flares, which look like lumps sometimes called gout under the skin, that do not attacks, per year go away when a flare stops that continues between flares To avoid gout and other problems, uric acid levels should be 6.0 mg/dL or below. If you have these signs and symptoms your uric acid level may need to be at or below 5 mg per dL. Take a Step in Controlling Gout 1 2 3 Learn more GoutRevealed.com See a gout specialist, most Talk about your gout symptoms, Visit the link above to hear from other commonly a rheumatologist. -
Arthritis (Overview)
ARTHRITIS Having arthritis can significantly affect your comfort & ability to walk and move with confidence. This is because it affects your joints, which are responsible for keeping you steady and moving efficiently. Your symptoms and causes will depend on the type of arthritis that you have. At Masterton Foot Clinic, our podiatrists work closely with patients with four types of arthritis. OSTEOARTHRITIS Osteoarthritis is the wear and tear arthritis that develops slowly over time as the cartilage that covers your bone ends wears down. The cause is largely from natural use over many years, though injuries, alignment issues within the joint and other diseases may result in it developing at a faster rate. We work with patients that want to feel more comfortable on their feet, despite having arthritis in their hip, knee, ankle and foot joints. RHEUMATOID ARTHRITIS Rheumatoid arthritis is an autoimmune disease that affects the joints. It occurs when your body’s immune system attacks the joints and causes damage, inflammation and pain. If the effects of rheumatoid arthritis remain uncontrolled, it can cause permanent changes in the appearance of the joints, especially at the feet and hands. We work with patients to help them manage the discomfort associated with rheumatoid arthritis, offloading prominent and painful areas that have developed due to changes in the joints. GOUT Gout is an inflammatory arthritis that results from a high concentration of uric acid in the blood. It is associated with a high intake of purine-containing foods like red meats, shellfish and red wine, hence it was previously referred to as the rich man’s disease. -

Table S1. List of Genes Up- Or Down-Regulated in H99 When Bound by 18B7
Table S1. List of genes up- or down-regulated in H99 when bound by 18B7. Up-regulated Genes Classification CNF03470: Formate dehydrogenase Metabolism CNC03960: Phosphate transporter, putative Metabolism CND00530: Putative urea transporter Secretion CNA02250: Ammonium transporter MEP1 Secretion CNC06440: Inositol 1-phosphate synthase Metabolism CND03490: Chitin deacetylase-like mannoprotein MP98 Cell wall CNA04560: Hypothetical protein Hypothetical CNF02180: Acetyl-CoA carboxylase, putative Metabolism CNJ00690: Uracil permease, putative Secretion CNF02510: Alcohol dehydrogenase, putative Metabolism CNE04360: Fatty acid synthase, alpha subunit-related Metabolism CNM00180: Cyclohydrolase, putative Metabolism CNL03740: AF540951 catalase isozyme P Stress CNE04370: Fatty acid synthase, beta subunit Metabolism CNA01790: Expressed protein Hypothetical CNA05700: Expressed protein Hypothetical CND03840: Vacuole fusion, non-autophagic-related protein, putative Cell wall CNM00980: Hypothetical protein Hypothetical CNI02420: Uricase (Urate oxidase), putative Metabolism CNJ01090: Xylitol dehydrogenase-related Cell wall CNC03430: Alpha-1,6-mannosyltransferase, putative Cell wall CNF04120: Expressed protein Hypothetical CNB01810: Short chain dehydrogenase, putative Metabolism CNJ01200: Hypothetical protein Hypothetical CNA01160: LSDR putative Metabolism CNH03430: Hypothetical protein Hypothetical CNM02410: Putative proteine disulfate isomerase Housekeeping CNG04200: Alpha-amylase, putative Cell wall CNC00920: NADP-dependent glutamate dehydrogenase Metabolism -

Nucleotide Metabolism 22
Nucleotide Metabolism 22 For additional ancillary materials related to this chapter, please visit thePoint. I. OVERVIEW Ribonucleoside and deoxyribonucleoside phosphates (nucleotides) are essential for all cells. Without them, neither ribonucleic acid (RNA) nor deoxyribonucleic acid (DNA) can be produced, and, therefore, proteins cannot be synthesized or cells proliferate. Nucleotides also serve as carriers of activated intermediates in the synthesis of some carbohydrates, lipids, and conjugated proteins (for example, uridine diphosphate [UDP]-glucose and cytidine diphosphate [CDP]- choline) and are structural components of several essential coenzymes, such as coenzyme A, flavin adenine dinucleotide (FAD[H2]), nicotinamide adenine dinucleotide (NAD[H]), and nicotinamide adenine dinucleotide phosphate (NADP[H]). Nucleotides, such as cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP), serve as second messengers in signal transduction pathways. In addition, nucleotides play an important role as energy sources in the cell. Finally, nucleotides are important regulatory compounds for many of the pathways of intermediary metabolism, inhibiting or activating key enzymes. The purine and pyrimidine bases found in nucleotides can be synthesized de novo or can be obtained through salvage pathways that allow the reuse of the preformed bases resulting from normal cell turnover. [Note: Little of the purines and pyrimidines supplied by diet is utilized and is degraded instead.] II. STRUCTURE Nucleotides are composed of a nitrogenous base; a pentose monosaccharide; and one, two, or three phosphate groups. The nitrogen-containing bases belong to two families of compounds: the purines and the pyrimidines. A. Purine and pyrimidine bases Both DNA and RNA contain the same purine bases: adenine (A) and guanine (G). -

Gout and Monoarthritis
Gout and Monoarthritis Acute monoarthritis has numerous causes, but most commonly is related to crystals (gout and pseudogout), trauma and infection. Early diagnosis is critical in order to identify and treat septic arthritis, which can lead to rapid joint destruction. Joint aspiration is the gold standard method of diagnosis. For many reasons, managing gout, both acutely and as a chronic disease, is challenging. Registrars need to develop a systematic approach to assessing monoarthritis, and be familiar with the management of gout and other crystal arthropathies. TEACHING AND • Aetiology of acute monoarthritis LEARNING AREAS • Risk factors for gout and septic arthritis • Clinical features and stages of gout • Investigation of monoarthritis (bloods, imaging, synovial fluid analysis) • Joint aspiration techniques • Interpretation of synovial fluid analysis • Management of hyperuricaemia and gout (acute and chronic), including indications and targets for urate-lowering therapy • Adverse effects of medications for gout, including Steven-Johnson syndrome • Indications and pathway for referral PRE- SESSION • Read the AAFP article - Diagnosing Acute Monoarthritis in Adults: A Practical Approach for the Family ACTIVITIES Physician TEACHING TIPS • Monoarthritis may be the first symptom of an inflammatory polyarthritis AND TRAPS • Consider gonococcal infection in younger patients with monoarthritis • Fever may be absent in patients with septic arthritis, and present in gout • Fleeting monoarthritis suggests gonococcal arthritis or rheumatic fever -

SUPPY Liglucosexlmtdh
US 20100314248A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2010/0314248 A1 Worden et al. (43) Pub. Date: Dec. 16, 2010 (54) RENEWABLE BOELECTRONIC INTERFACE Publication Classification FOR ELECTROBOCATALYTC REACTOR (51) Int. Cl. (76) Inventors: Robert M. Worden, Holt, MI (US); C25B II/06 (2006.01) Brian L. Hassler, Lake Orion, MI C25B II/2 (2006.01) (US); Lawrence T. Drzal, Okemos, GOIN 27/327 (2006.01) MI (US); Ilsoon Lee, Okemo s, MI BSD L/04 (2006.01) (US) C25B 9/00 (2006.01) (52) U.S. Cl. ............... 204/403.14; 204/290.11; 204/400; Correspondence Address: 204/290.07; 427/458; 204/252: 977/734; PRICE HENEVELD COOPER DEWITT & LIT 977/742 TON, LLP 695 KENMOOR, S.E., PO BOX 2567 (57) ABSTRACT GRAND RAPIDS, MI 495.01 (US) An inexpensive, easily renewable bioelectronic device useful for bioreactors, biosensors, and biofuel cells includes an elec (21) Appl. No.: 12/766,169 trically conductive carbon electrode and a bioelectronic inter face bonded to a surface of the electrically conductive carbon (22) Filed: Apr. 23, 2010 electrode, wherein the bioelectronic interface includes cata lytically active material that is electrostatically bound directly Related U.S. Application Data or indirectly to the electrically conductive carbon electrode to (60) Provisional application No. 61/172,337, filed on Apr. facilitate easy removal upon a change in pH, thereby allowing 24, 2009. easy regeneration of the bioelectronic interface. 7\ POWER 1 - SUPPY|- LIGLUCOSEXLMtDH?till pi 6.0 - esses&aaaas-exx-xx-xx-xx-xxxxixax-e- Patent Application Publication Dec. 16, 2010 Sheet 1 of 18 US 2010/0314248 A1 Potential (nV) Patent Application Publication Dec. -

Monitoring the Redox Status in Multiple Sclerosis
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 31 July 2020 doi:10.20944/preprints202007.0737.v1 Review Monitoring the Redox Status in Multiple Sclerosis Masaru Tanaka 1,2 and László Vécsei 1,2,* 1 MTA-SZTE, Neuroscience Research Group, Semmelweis u. 6, Szeged, H-6725 Hungary; [email protected] 2 Department of Neurology, Interdisciplinary Excellence Centre, Faculty of Medicine, University of Szeged, Semmelweis u. 6, H-6725 Szeged, Hungary * Correspondence: [email protected]; Tel.: +36-62-545-351 Received: date; Accepted: date; Published: date Abstract: Worldwide, over 2.2 million people are suffered from multiple sclerosis (MS), a multifactorial demyelinating disease of the central nervous system, characterized by multifocal inflammatory or demyelinating attacks associated with neuroinflammation and neurodegeneration. The blood, cerebrospinal fluid, and postmortem brain samples of MS patients evidenced the presence of reduction-oxidation (redox) homeostasis disturbance such as the alternations of oxidative and antioxidative enzyme activities and the presence of degradation products. This review article discussed the components of redox homeostasis including reactive chemical species, oxidative enzymes, antioxidative enzymes, and degradation products. The reactive chemical species covered frequently discussed reactive oxygen/nitrogen species, rarely featured reactive chemicals such as sulfur, carbonyls, halogens, selenium, and nucleophilic species that potentially act as reductive as well as pro-oxidative stressors. The antioxidative enzyme systems covered the nuclear factor erythroid-2-related factor 2 (NRF2)-Kelch-like ECH-associated protein 1 (KEAP1) signaling pathway, a possible biomarker sensitive to the initial phase of oxidative stress. Altered components of the redox homeostasis in MS were discussed, some of which turned to be MS subtype- or treatment-specific and thus potentially become diagnostic, prognostic, predictive, and/or therapeutic biomarkers. -

19120 Arthritis Aus Gout Booklet
Taking control of your Gout A practical guide to treatments, services and lifestyle choices How can this booklet help you This booklet is designed for people who have gout. It will help you understand your • make healthy choices for your condition so that you can better general health and wellbeing manage your symptoms and continue • find support and additional to lead an active and healthy life. information to cope with the This booklet offers information and impact of gout. practical advice to help you: The information inside is based • understand what gout is and on the latest research and what it means for you recommendations, and has been reviewed by Australian experts in the • understand how medicines can field of arthritis to make sure it is help treat gout attacks and current and relevant to your needs. prevent future attacks So go ahead — take control of • work with your healthcare team your gout! to manage the disease in the short and long term © Copyright Arthritis Australia 2014 Supported by: AstraZeneca Pty Limited ABN 54 009 682 311 Alma Road, North Ryde NSW 2113 2 Taking control of your Gout Contents Understanding gout 4 Treating gout 10 Diet and lifestyle 16 Who can help? 21 Working with your GP 22 Seeing a rheumatologist 23 Other health professionals 24 Seeking support 26 Glossary of terms 28 Useful resources 29 Medical and consumer consultants Tanya deKroo, Information Resources Coordinator, Arthritis Australia Wendy Favorito, Arthritis Australia Consumer Representative and Board Member Assoc Prof Neil McGill, Rheumatologist Assoc Prof Julian McNeil, Rheumatologist and Chair of Australian Rheumatology Association’s Therapeutics Committee Assoc Prof Peter Youssef, Rheumatologist and Chair of Arthritis Australia’s Scientific Advisory Committee Arthritis Australia 3 Understanding gout What is gout? the main reason for more than Gout is an extremely painful form nine out of ten people with gout). -

21362 Arthritis Australia a to Z List
ARTHRITISINFORMATION SHEET Here is the A to Z of arthritis! A D Goodpasture’s syndrome Achilles tendonitis Degenerative joint disease Gout Achondroplasia Dermatomyositis Granulomatous arteritis Acromegalic arthropathy Diabetic finger sclerosis Adhesive capsulitis Diffuse idiopathic skeletal H Adult onset Still’s disease hyperostosis (DISH) Hemarthrosis Ankylosing spondylitis Discitis Hemochromatosis Anserine bursitis Discoid lupus erythematosus Henoch-Schonlein purpura Avascular necrosis Drug-induced lupus Hepatitis B surface antigen disease Duchenne’s muscular dystrophy Hip dysplasia B Dupuytren’s contracture Hurler syndrome Behcet’s syndrome Hypermobility syndrome Bicipital tendonitis E Hypersensitivity vasculitis Blount’s disease Ehlers-Danlos syndrome Hypertrophic osteoarthropathy Brucellar spondylitis Enteropathic arthritis Bursitis Epicondylitis I Erosive inflammatory osteoarthritis Immune complex disease C Exercise-induced compartment Impingement syndrome Calcaneal bursitis syndrome Calcium pyrophosphate dehydrate J (CPPD) F Jaccoud’s arthropathy Crystal deposition disease Fabry’s disease Juvenile ankylosing spondylitis Caplan’s syndrome Familial Mediterranean fever Juvenile dermatomyositis Carpal tunnel syndrome Farber’s lipogranulomatosis Juvenile rheumatoid arthritis Chondrocalcinosis Felty’s syndrome Chondromalacia patellae Fibromyalgia K Chronic synovitis Fifth’s disease Kawasaki disease Chronic recurrent multifocal Flat feet Kienbock’s disease osteomyelitis Foreign body synovitis Churg-Strauss syndrome Freiberg’s disease -
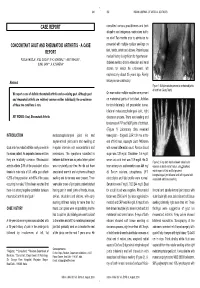
CONCOMITANT GOUT and RHEUMATOID ARTHRITIS - a CASE Presented with Multiple Nodular Swellings on REPORT Feet, Hands, Wrists and Elbows
349 350 INDIAN JOURNAL OF MEDICAL SCIENCES CASE REPORT consulted various practitioners and took allopathic and indigenous medications but to no relief. Two months prior to admission he CONCOMITANT GOUT AND RHEUMATOID ARTHRITIS - A CASE presented with multiple nodular swellings on REPORT feet, hands, wrists and elbows. Patient’s past medical history is significant for hypertension, POOJA KHOSLA*, ATUL GOGIA**, P. K. AGARWAL***, AMIT PAHUJA**, SUNIL JAIN***, K. K. SAXENA# diabetes mellitus, chronic ethanolism and renal stones for which he underwent left nephrectomy about 25 years ago. Family Abstract history is non-contributory. Figure 1: Multiple nodules present on metatarsal joints of both feet (Gouty Tophi) We report a case of definite rheumatoid arthritis and co-existing gout. Although gout On examination multiple nodules were present and rheumatoid arthritis are relatively common entities individually, the co-existence on metatarsal joints of both feet, Achilles of these two conditions is rare. tendon bilaterally, left prepatellar bursa, bilateral metacarpophalangeal joint, right KEY WORDS: Gout, Rheumatoid Arthritis olecranon process. There was swelling and tenderness of PIP and MCP joints of both feet. (Figure 1) Laboratory data revealed INTRODUCTION metatarsophalangeal joint. He had hemoglobin - 8.9gm/dl, ESR 124 mm at the asymmetrical joint pains and swelling at end of first hour, leucocyte count 7400/cmm Gout and rheumatoid arthritis rarely co-exist in irregular intervals with exacerbations and with normal differential count. Random blood the same patient. As separate disease entities remissions. The symptoms subsided in sugar was 139 mg/dl, Creatinine- 1.6 mg/dl; they are relatively common. Rheumatoid between but there was no period when patient serum uric acid level was 10.9 mg/dl. -

Psoriasis, Psoriatic Arthritis and Secondary Gout – 3 Case Reports
https://doi.org/10.5272/jimab.2018242.1972 Journal of IMAB Journal of IMAB - Annual Proceeding (Scientific Papers). 2018 Apr-Jun;24(2) ISSN: 1312-773X https://www.journal-imab-bg.org Case report PSORIASIS, PSORIATIC ARTHRITIS AND SECONDARY GOUT – 3 CASE REPORTS Mina I. Ivanova, Rositsa Karalilova, Zguro Batalov. Departement of Rheumatology, Faculty of Medicine, UMHAT Kaspela, Medical University - Plovdiv, Bulgaria. ABSTRACT: cells in the skin (Langerhans cells) migrate to the regional Background: One of the most common complica- lymph nodes, where interacts with T-lymphocytes. This tions in psoriasis is the development of secondary gout that provokes the immune response leading to the activation often remains undiagnosed for many years. In some cases, of T cells and release of cytokines. The local effects of the clinical symptoms of gout precede the manifestation cytokines lead to a cell-mediated immune response. In pso- of cutaneous psoriasis, leading to progression of the dis- riasis skin lesions are results of hyperplasia of the epider- ease and early onset of complications. According to the mis, which leads to enhanced cell reproduction, world data, there is a strong correlation between psoriasis hyperproliferation and at the same time shortening vulgaris and psoriatic arthritis on the one hand and gout keratinocytes’ life. This leads to increased production of on the other ranging from 3 to 40%. In Bulgaria, there are uric acid, as it enhances the exchange of nucleic acids. no studies observing the frequency of secondary gout in Psoriatic arthritis (PsA) occurs in 0.05 to 0.25% from psoriatic patients. the general population.