Mouse Pphln1 Conditional Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Genome-Wide DNA Methylation Profiling Identifies Differential Methylation in Uninvolved Psoriatic Epidermis
Genome-Wide DNA Methylation Profiling Identifies Differential Methylation in Uninvolved Psoriatic Epidermis Deepti Verma, Anna-Karin Ekman, Cecilia Bivik Eding and Charlotta Enerbäck The self-archived postprint version of this journal article is available at Linköping University Institutional Repository (DiVA): http://urn.kb.se/resolve?urn=urn:nbn:se:liu:diva-147791 N.B.: When citing this work, cite the original publication. Verma, D., Ekman, A., Bivik Eding, C., Enerbäck, C., (2018), Genome-Wide DNA Methylation Profiling Identifies Differential Methylation in Uninvolved Psoriatic Epidermis, Journal of Investigative Dermatology, 138(5), 1088-1093. https://doi.org/10.1016/j.jid.2017.11.036 Original publication available at: https://doi.org/10.1016/j.jid.2017.11.036 Copyright: Elsevier http://www.elsevier.com/ Genome-Wide DNA Methylation Profiling Identifies Differential Methylation in Uninvolved Psoriatic Epidermis Deepti Verma*a, Anna-Karin Ekman*a, Cecilia Bivik Edinga and Charlotta Enerbäcka *Authors contributed equally aIngrid Asp Psoriasis Research Center, Department of Clinical and Experimental Medicine, Division of Dermatology, Linköping University, Linköping, Sweden Corresponding author: Charlotta Enerbäck Ingrid Asp Psoriasis Research Center, Department of Clinical and Experimental Medicine, Linköping University SE-581 85 Linköping, Sweden Phone: +46 10 103 7429 E-mail: [email protected] Short title Differential methylation in psoriasis Abbreviations CGI, CpG island; DMS, differentially methylated site; RRBS, reduced representation bisulphite sequencing Keywords (max 6) psoriasis, epidermis, methylation, Wnt, susceptibility, expression 1 ABSTRACT Psoriasis is a chronic inflammatory skin disease with both local and systemic components. Genome-wide approaches have identified more than 60 psoriasis-susceptibility loci, but genes are estimated to explain only one third of the heritability in psoriasis, suggesting additional, yet unidentified, sources of heritability. -

TASOR Is a Pseudo-PARP That Directs HUSH Complex Assembly and Epigenetic Transposon Control
Lawrence Berkeley National Laboratory Recent Work Title TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Permalink https://escholarship.org/uc/item/6021r2cd Journal Nature communications, 11(1) ISSN 2041-1723 Authors Douse, Christopher H Tchasovnikarova, Iva A Timms, Richard T et al. Publication Date 2020-10-02 DOI 10.1038/s41467-020-18761-6 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ARTICLE https://doi.org/10.1038/s41467-020-18761-6 OPEN TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control Christopher H. Douse 1,4,9, Iva A. Tchasovnikarova2,5,9, Richard T. Timms 2,9, Anna V. Protasio 2,6, Marta Seczynska2, Daniil M. Prigozhin 1,7, Anna Albecka1,2,8, Jane Wagstaff3, James C. Williamson2, ✉ ✉ Stefan M. V. Freund3, Paul J. Lehner 2 & Yorgo Modis 1,2 1234567890():,; The HUSH complex represses retroviruses, transposons and genes to maintain the integrity of vertebrate genomes. HUSH regulates deposition of the epigenetic mark H3K9me3, but how its three core subunits — TASOR, MPP8 and Periphilin — contribute to assembly and targeting of the complex remains unknown. Here, we define the biochemical basis of HUSH assembly and find that its modular architecture resembles the yeast RNA-induced tran- scriptional silencing complex. TASOR, the central HUSH subunit, associates with RNA pro- cessing components. TASOR is required for H3K9me3 deposition over LINE-1 repeats and repetitive exons in transcribed genes. In the context of previous studies, this suggests that an RNA intermediate is important for HUSH activity. We dissect the TASOR and MPP8 domains necessary for transgene repression. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Download Author Version (PDF)
Molecular BioSystems Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/molecularbiosystems Page 1 of 29 Molecular BioSystems Mutated Genes and Driver Pathways Involved in Myelodysplastic Syndromes—A Transcriptome Sequencing Based Approach Liang Liu1*, Hongyan Wang1*, Jianguo Wen2*, Chih-En Tseng2,3*, Youli Zu2, Chung-che Chang4§, Xiaobo Zhou1§ 1 Center for Bioinformatics and Systems Biology, Division of Radiologic Sciences, Wake Forest University Baptist Medical Center, Winston-Salem, NC 27157, USA. 2 Department of Pathology, the Methodist Hospital Research Institute, -

Oncogenic Fusion Protein FGFR2-PPHLN1: Requirements for Biological Activation, and Efficacy of Inhibitors
UC San Diego UC San Diego Previously Published Works Title Oncogenic fusion protein FGFR2-PPHLN1: Requirements for biological activation, and efficacy of inhibitors. Permalink https://escholarship.org/uc/item/12g996rt Journal Translational oncology, 13(12) ISSN 1936-5233 Authors Li, Fangda Meyer, April N Peiris, Malalage N et al. Publication Date 2020-12-01 DOI 10.1016/j.tranon.2020.100853 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Translational Oncology 13 (2020) 100853 Contents lists available at ScienceDirect Translational Oncology journal homepage: www.elsevier.com/locate/tranon Oncogenic fusion protein FGFR2-PPHLN1: Requirements for biological activation, and efficacy of inhibitors ⁎ Fangda Li a, April N. Meyer a, Malalage N. Peiris a, Katelyn N. Nelson a, Daniel J. Donoghue a,b, a Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA 92093-0367, USA b UCSD Moores Cancer Center and Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA 92093-0367, USA ARTICLE INFO ABSTRACT Article history: Aim of study: Chromosomal translocations such as t(10;12)(q26,q12) are associated with intrahepatic cholangiocarci- Received 2 May 2020 noma, a universally fatal category of liver cancer. This translocation creates the oncogenic fusion protein of Fibroblast Received in revised form 24 July 2020 Growth Factor Receptor 2 joined to Periphilin 1. The aims of this study were to identify significant features required for Accepted 9 August 2020 biological activation, analyze the activation of downstream signaling pathways, and examine the efficacy of the TKIs Available online xxxx BGJ398 and TAS-120, and of the MEK inhibitor Trametinib. -

PPHLN1 Rabbit Pab
Leader in Biomolecular Solutions for Life Science PPHLN1 Rabbit pAb Catalog No.: A17706 Basic Information Background Catalog No. The protein encoded by this gene is one of the several proteins that become sequentially A17706 incorporated into the cornified cell envelope during the terminal differentiation of keratinocyte at the outer layers of epidermis. This protein interacts with periplakin, Observed MW which is known as a precursor of the cornified cell envelope. The cellular localization 53kDa pattern and insolubility of this protein suggest that it may play a role in epithelial differentiation and contribute to epidermal integrity and barrier formation. Multiple Calculated MW alternatively spliced transcript variants encoding distinct isoforms have been observed. [provided by RefSeq, Jul 2008] Category Primary antibody Applications WB Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 51535 Q8NEY8 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 195-255 of human PPHLN1 (NP_958846.1). Synonyms CR;HSPC206;HSPC232;PPHLN1 Contact Product Information 400-999-6126 Source Isotype Purification Rabbit IgG Affinity purification [email protected] www.abclonal.com.cn Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using PPHLN1 antibody (A17706) at 1:1000 dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) (AS014) at 1:10000 dilution. Lysates/proteins: 25ug per lane. Blocking buffer: 3% nonfat dry milk in TBST. Detection: ECL Basic Kit (RM00020). Exposure time: 10s. Antibody | Protein | ELISA Kits | Enzyme | NGS | Service For research use only. -

Oncogenic Fusion Protein FGFR2-PPHLN1: Requirements for Biological Activation, and Efficacy of Inhibitors ⁎ Fangda Li A, April N
Translational Oncology 13 (2020) 100853 Contents lists available at ScienceDirect Translational Oncology journal homepage: www.elsevier.com/locate/tranon Oncogenic fusion protein FGFR2-PPHLN1: Requirements for biological activation, and efficacy of inhibitors ⁎ Fangda Li a, April N. Meyer a, Malalage N. Peiris a, Katelyn N. Nelson a, Daniel J. Donoghue a,b, a Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA 92093-0367, USA b UCSD Moores Cancer Center and Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA 92093-0367, USA ARTICLE INFO ABSTRACT Article history: Aim of study: Chromosomal translocations such as t(10;12)(q26,q12) are associated with intrahepatic cholangiocarci- Received 2 May 2020 noma, a universally fatal category of liver cancer. This translocation creates the oncogenic fusion protein of Fibroblast Received in revised form 24 July 2020 Growth Factor Receptor 2 joined to Periphilin 1. The aims of this study were to identify significant features required for Accepted 9 August 2020 biological activation, analyze the activation of downstream signaling pathways, and examine the efficacy of the TKIs Available online xxxx BGJ398 and TAS-120, and of the MEK inhibitor Trametinib. Methods: These studies examined FGFR2-PPHLN1 proteins containing a kinase-dead, kinase-activated, or WT kinase domain in comparison with analogous FGFR2 proteins. Biological activity was assayed using soft agar colony forma- tion in epithelial RIE-1 cells and focus assays in NIH3T3 cells. The MAPK/ERK, JAK/STAT3 and PI3K/AKT signaling pathways were examined for activation. Membrane association was analyzed by indirect immunofluorescence com- paring proteins altered by deletion of the signal peptide, or by addition of a myristylation signal. -

FGF/FGFR Signaling in Health and Disease
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN FGF/FGFR signaling in health and disease Yangli Xie1, Nan Su1, Jing Yang1, Qiaoyan Tan1, Shuo Huang 1, Min Jin1, Zhenhong Ni1, Bin Zhang1, Dali Zhang1, Fengtao Luo1, Hangang Chen1, Xianding Sun1, Jian Q. Feng2, Huabing Qi1 and Lin Chen 1 Growing evidences suggest that the fibroblast growth factor/FGF receptor (FGF/FGFR) signaling has crucial roles in a multitude of processes during embryonic development and adult homeostasis by regulating cellular lineage commitment, differentiation, proliferation, and apoptosis of various types of cells. In this review, we provide a comprehensive overview of the current understanding of FGF signaling and its roles in organ development, injury repair, and the pathophysiology of spectrum of diseases, which is a consequence of FGF signaling dysregulation, including cancers and chronic kidney disease (CKD). In this context, the agonists and antagonists for FGF-FGFRs might have therapeutic benefits in multiple systems. Signal Transduction and Targeted Therapy (2020) 5:181; https://doi.org/10.1038/s41392-020-00222-7 INTRODUCTION OF THE FGF/FGFR SIGNALING The binding of FGFs to the inactive monomeric FGFRs will Fibroblast growth factors (FGFs) are broad-spectrum mitogens and trigger the conformational changes of FGFRs, resulting in 1234567890();,: regulate a wide range of cellular functions, including migration, dimerization and activation of the cytosolic tyrosine kinases by proliferation, differentiation, and survival. It is well documented phosphorylating the tyrosine residues within the cytosolic tail of that FGF signaling plays essential roles in development, metabo- FGFRs.4 Then, the phosphorylated tyrosine residues serve as the lism, and tissue homeostasis. -
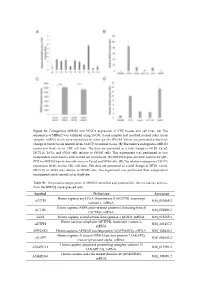
Figure S1. Endogenous MIR45
Figure S1. Endogenous MIR452 and VEGFA expression in CRC tissues and cell lines. (A) The expression of MIR452 was validated using 10 CRC tissue samples and matched normal colon tissue samples. miRNA levels were normalized to colon-specific RNU48. Values are presented as the fold- change in tumor tissue relative levels (ΔΔCT) to normal tissue. (B) The relative endogenous MIR452 expression levels in six CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, LoVo, and SW48 cells relative to SW480 cells. This experiment was performed as two independent experiments, each carried out in triplicate. (C) MIR452 expression level analysis by qRT- PCR for MIR452 transfection efficiency in Caco2 and SW48 cells. (D) The relative endogenous VEGFA expression levels in five CRC cell lines. The data are presented as a fold change in HT29, Caco2, HCT116, or SW48 cells relative to SW480 cells. This experiment was performed three independent experiments, each carried out in duplicate. Table S1. The putative target genes of MIR452 identified and predicted by the microarray analysis from the MIR452 overexpressed cells. Symbol Definition Accession Homo sapiens acyl-CoA thioesterase 8 (ACOT8), transcript ACOT8 NM_005469.2 variant 1, mRNA. Homo sapiens ARP6 actin-related protein 6 homolog (yeast) ACTR6 NM_022496.3 (ACTR6), mRNA. ADI1 Homo sapiens acireductone dioxygenase 1 (ADI1), mRNA. NM_018269.1 Homo sapiens aftiphilin (AFTPH), transcript variant 1, AFTPH NM_203437.2 mRNA. AHNAK2 Homo sapiens AHNAK nucleoprotein 2 (AHNAK2), mRNA. NM_138420.2 Homo sapiens A kinase (PRKA) anchor protein 7 (AKAP7), AKAP7 NM_004842.2 transcript variant alpha, mRNA. Homo sapiens anaphase promoting complex subunit 13 ANAPC13 NM_015391.2 (ANAPC13), mRNA. -

The HUSH Complex Cooperates with TRIM28 to Repress Young Retrotransposons and New Genes
Downloaded from genome.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Research The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes Luisa Robbez-Masson,1 Christopher H.C. Tie,1 Lucia Conde,2 Hale Tunbak,1 Connor Husovsky,1 Iva A. Tchasovnikarova,3 Richard T. Timms,3 Javier Herrero,2 Paul J. Lehner,3 and Helen M. Rowe1 1Infection and Immunity, University College London, London WC1E 6BT, United Kingdom; 2Bill Lyons Informatics Centre, UCL Cancer Institute, University College London, London WC1E 6DD, United Kingdom; 3Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, United Kingdom Retrotransposons encompass half of the human genome and contribute to the formation of heterochromatin, which pro- vides nuclear structure and regulates gene expression. Here, we asked if the human silencing hub (HUSH) complex is nec- essary to silence retrotransposons and whether it collaborates with TRIM28 and the chromatin remodeler ATRX at specific genomic loci. We show that the HUSH complex contributes to de novo repression and DNA methylation of an SVA retro- transposon reporter. By using naıvë versus primed mouse pluripotent stem cells, we reveal a critical role for the HUSH com- plex in naıvë cells, implicating it in programming epigenetic marks in development. Although the HUSH component FAM208A binds to endogenous retroviruses (ERVs) and long interspersed element-1s (LINE-1s or L1s), it is mainly required to repress evolutionarily young L1s (mouse-specific lineages <5 million years old). TRIM28, in contrast, is necessary to repress both ERVs and young L1s. Genes co-repressed by TRIM28 and FAM208A are evolutionarily young, or exhibit tissue-specific expression, are enriched in young L1s, and display evidence for regulation through LTR promoters.