Cannabinoid As Potential Aromatase Inhibitor Through Molecular Modeling and Screening for Anti-Cancer Activity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Targeting the Endocannabinoid System to Reduce Nociception
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2011 Targeting the endocannabinoid system to reduce nociception Lamont Booker Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Medical Pharmacology Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/2419 This Dissertation is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Targeting the Endocannabinoid System to Reduce Nociception A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy at Virginia Commonwealth University. By Lamont Booker Bachelor’s of Science, Fayetteville State University 2003 Master’s of Toxicology, North Carolina State University 2005 Director: Dr. Aron H. Lichtman, Professor, Pharmacology & Toxicology Virginia Commonwealth University Richmond, Virginia April 2011 Acknowledgements The author wishes to thank several people. I like to thank my advisor Dr. Aron Lichtman for taking a chance and allowing me to work under his guidance. He has been a great influence not only with project and research direction, but as an excellent example of what a mentor should be (always willing to listen, understanding the needs of each student/technician, and willing to provide a hand when available). Additionally, I like to thank all of my committee members (Drs. Galya Abdrakmanova, Francine Cabral, Sandra Welch, Mike Grotewiel) for your patience and willingness to participate as a member. Our term together has truly been memorable! I owe a special thanks to Sheryol Cox, and Dr. -

CBD (Cannabidiol)
TRANSPORTATION RESEARCH BOARD Driving Toward the Truth - Dispelling the Myths About Cannabis Products February 10, 2021 @NASEMTRB #TRBwebinar PDH Certification The Transportation Research Board has met the standards and Information: requirements of the Registered Continuing Education Providers •1.5 Professional Development Program. Credit earned on completion Hour (PDH) – see follow-up of this program will be reported to email for instructions RCEP. A certificate of completion will •You must attend the entire be issued to participants that have registered and attended the entire webinar to be eligible to receive session. As such, it does not include PDH credits content that may be deemed or •Questions? Contact Reggie construed to be an approval or Gillum at [email protected] endorsement by RCEP. #TRBwebinar Learning Objectives 1. Identify impacts of the Farm Bill on use of THC and CBD products 2. Describe the toxicology of THC and CBD products 3. Discuss how THC and CBD products affect driving performance and crash risk #TRBwebinar TRB Standing Committee on Impairment in Transportation (ACS50) TRB Webinar: Driving Toward the Truth - Dispelling the Myths About Cannabis Products Dr. Barry K. Logan Executive Director, Center for Forensic Science Research and Education (CFSRE); Senior Vice President of Forensic Sciences, and Chief Scientist at NMS Labs Michelle Peace, Ph.D. Associate Professor and PI, Laboratory for Forensic Toxicology Research Department of Forensic Science, Virginia Commonwealth University Dr. Darrin Grondel Vice President, -

Medical Marijuana: High Expectations
Medical Marijuana: High Expectations Dr. Ally Dering-Anderson, RP University of Nebraska College of Pharmacy August 2019 What we know What we don’t Speaker Contact Information Dr. Ally Dering-Anderson, RP Clinical Associate Professor of Pharmacy University of Nebraska College of Pharmacy 986145 Medical Center College of Pharmacy Department of Pharmacy Practice Omaha, NE 68198-6145 [email protected] Speaker Disclosures Dr. Ally is on the faculty of the University of Nebraska College of Pharmacy Department of Pharmacy Practice She is the Senior Partner of BBfN, a health and safety consulting company She has no conflict in regard to medical use of cannabinoids “Off Label Use” In order to assure that there are no problems with the accreditation of this program nor problems caused for the accrediting body here goes: There are 3 approved products containing one or more chemicals found in the marijuana plant. Epidiolex (cannabidiol) Marinol Cesamet We will NOT discuss uses for these 3 approved products beyond the labeling. We will discuss cannabinoids not found in drug products currently approved by the FDA. The concept of being “off label” for these products is illogical, as there is no label to follow. Got it? Bibliography: #’s 1, 2, 3 A note from the CE clinical reviewers … “Medical marijuana is considered a controversial topic in CME. When a CME activity includes information about an approach to diagnosis or treatment that is not generally accepted, it is allowable to facilitate debate and discussion about the approach, but it is not allowable to advocate for the test or treatment, or teach clinicians how or when to use it.” Trust me, I am not advocating for any test - - in fact, I’m going to slam one. -

Crosstalk Between Cannabinoid Receptors and Epidermal
CROSSTALK BETWEEN CANNABINOID RECEPTORS AND EPIDERMAL GROWTH FACTOR RECEPTOR IN NON-SMALL CELL LUNG CANCER DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By JANANI RAVI Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2015 Dissertation Committee: Dr. Ramesh Ganju, Advisor Dr. Kalpana Ghoshal Dr. Xianghong Zou Dr. Sujit Basu Copyright by Janani Ravi 2015 Abstract (i) The endocannabinoid anandamide (AEA), a neurotransmitter was shown to have anti- cancer effects. Fatty acid amide hydrolase (FAAH) metabolizes AEA and decreases its anti-tumorigenic activity. In this study, we have analyzed the role of FAAH inhibition in non-small cell lung cancer (NSCLC). We have shown that FAAH and CB1 receptor which is activated by AEA are expressed in lung adenocarcinoma patient samples and NSCLC cell lines A549 and H460. Since the synthetic analogue of anandamide (Met-F- AEA) did not possess significant anti-tumorigenic effects, we used Met-F-AEA in combination with FAAH inhibitor URB597 which significantly reduced EGF (epidermal growth factor)-induced proliferative and chemotactic activities in vitro when compared to anti-tumorigenic activity of Met-F-AEA alone. Further analysis of signaling mechanisms revealed that Met-F-AEA in combination with URB597 inhibits activation of EGFR and its downstream signaling ERK, AKT and NF-kB. In addition, it inhibited MMP2 secretion and stress fiber formation. We have also shown that the Met-F-AEA in combination with URB597 induces G0/G1 cell cycle arrest by downregulating cyclin D1 and CDK4 expressions, ultimately leading to apoptosis via activation of caspase-9 and PARP. -

What Is Delta-8 THC?? Cannabinoid Chemistry 101
What is Delta-8 THC?? Cannabinoid Chemistry 101 National Conference on Weights and Measures Annual Meeting - Rochester, NY Matthew D. Curran, Ph.D. July 21, 2021 Disclaimer Just to be clear… • I am a chemist and not a lawyer so: • This presentation will not discuss the legal aspects of Δ8-THC or DEA’s current position. • This presentation will not discuss whether Δ8-THC is considered “synthetic” or “naturally occurring.” • This is not a position statement on any issues before the NCWM. • Lastly, this should only be considered a scientific sharing exercise. Florida Department of Agriculture and Consumer Services 2 Cannabis in Florida Cannabis Syllabus • What is Cannabis? • “Mother” Cannabinoid • Decarboxylation • Relationship between CBD and THC • What does “Total” mean? • Dry Weight vs. Wet Weight • What does “Delta-9” mean? • Relationship between “Delta-8” and “Delta-9” • CBD to Delta-8 THC • Cannabinoid Chemistry 202… Florida Department of Agriculture and Consumer Services 3 Cannabis Cannabis • Cannabis sativa is the taxonomic name for the plant. • The concentration of Total Δ9-Tetrahydrocannabinol (Total Δ9-THC) is critical when considering the varieties of Cannabis sativa. • Hemp – (Total Δ9-THC) 0.3% or less • Not really a controversial term, “hemp” • Marijuana/cannabis – (Total Δ9-THC) Greater than 0.3% • Controversial term, “marijuana” • Some states prohibit the use of this term whereas some states have it in their laws. • Some states use the term “cannabis.” • Not italicized • Lower case “c” Florida Department of Agriculture and -

Roles of Cannabidiol in Reversing Proteinopathies
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 21 May 2020 doi:10.20944/preprints202005.0340.v1 Review Roles of Cannabidiol in Reversing Proteinopathies Raju Dash1, Md. Chayan Ali2†, Israt Jahan3†, Yeasmin Akter Munni1†, Sarmistha Mitra4, Md. Abdul Hannan1,5, Binod Timalsina1, Diyah Fatimah Oktaviani1, Ho Jin Choi1, Il Soo Moon1* 1Department of Anatomy, Dongguk University College of Medicine, Gyeongju 38066, Republic of Korea 2Department of Biotechnology and Genetic Engineering, Faculty of Biological Sciences, Islamic University, Kushtia 7003, Bangladesh 3Department of Pharmacy, Faculty of Life and Earth Sciences, Jagannath University, Dhaka 1100, Bangladesh. 4Plasma Bioscience Research Center and Department of Plasma Bio display, Kwangwoon University, Seoul 01897, Republic of Korea 5Department of Biochemistry and Molecular Biology, Bangladesh Agricultural University, Mymensingh 2202, Bangladesh †These authors have contributed equally. *Corresponding author: Il Soo Moon Mailing address: Department of Anatomy, Dongguk University College of Medicine, 123 Dongdae-ro, Gyeongju 38066, Republic of Korea Phone: +82-54-770-2414 Fax: +82-54-770-2447 Email: [email protected] Abstract: Cannabidiol is a well-known non-psychotropic phytocannabinoid from Cannabis sativa, which exerts a broad range of neuropharmacological activities in the central nervous systems. Over the past years, compelling evidence from preclinical and clinical studies support therapeutic potentials of cannabidiol in various neurological disorders, including neurodegenerative diseases. Neurodegenerative diseases are characterized by the accumulation of misfolded or aggregated protein due to the defective protein homeostasis or proteostasis network, termed as proteinopathies. Because of its role in the protein homeostasis network, cannabidiol could be a potent molecule to revert not only age-associated neurodegeneration but also other protein misfolding disorders. -

In Silico Assessment of Drug-Like Properties of Phytocannabinoids in Cannabis Sativa
EDUCATUM JSMT Vol. 4 No. 2 (2017) ISSN 2289-7070 / eISSN 2462-2451 (1-7) https://ejournal.upsi.edu.my/journal/EDSC In Silico Assessment of Drug-Like Properties of Phytocannabinoids in Cannabis Sativa Shakinaz Desa1*, Asiah Osman2, and Richard Hyslop3 1Department of Biology, Universiti Pendidikan Sultan Idris, Malaysia, 2Natural Product Division, Forest Research Institute Malaysia, 3Department of Chemistry and Biochemistry, University of Northern Colorado, USA *Corresponding author: [email protected] Abstract This study investigated drug-like properties of phytocannabinoids in Cannabis sativa using an in silico study. We report sixteen phytocannabinoids: cannabidiol (CBD), cannabidiolic acid (CBDA), cannabinol (CBN), cannabichromene (CBC), cannabigerol (CBG), cannabicyclol (CBL), cannabivarin (CBV), cannabidivarin (CBDV), cannabichromevarin (CBCV), cannabigerovarin (CBGV), cannabinodiol (CBDL), cannabielsoin (CBE), cannabitriol (CBT), Δ9-tetrahydrocannabinol (Δ9-THC), Δ9-tetrahydrocannabivarin (Δ9-THCV), and Δ8-tetrahydrocannabinol (Δ8-THC). All chemical structures and properties were obtained from PubChem Compound, National Center for Biotechnology Information, U.S. National Library of Medicine. Molinspiration was used for the calculation of molecular properties and bioactivity score. The parameters were molecular weight (MW), number of hydrogen acceptor (HBA), number of hydrogen donor (HBD), partition coefficient (cLogP), polar surface area (PSA) and number of rotatable bonds (NROTB). We predicted bioactivity scores for G Protein-Coupled Receptors (GPCR) ligand, ion channel modulator, kinase inhibitor, nuclear receptor ligand, protease inhibitor and enzyme inhibitor. Lipinski’s rule was used as reference to determine the drug-like properties of the phytocannabinoids. All compounds have MW<500, HBA<10, HBD<5, TPSA<140Å2 and NRTOB<10. Bioactivity score showed an active or moderately active in all compounds. -

2 Cannabis Sativa L
CANNABIS SATIVA 29 2 Cannabis sativa L. Common Names Almindelig hamp Denmark Harilik kanep Slovenia Asa Japan Haschischpflanze Germany Bang Egypt Hash United Kingdom Bhaang India Hashas Turkey Bhaango Nepal Hashish Morocco Canamo indico Spain Hemp United Kingdom Canapa indica Italy Hennep Netherlands Canhamo Portugal Hind kinnabi Turkey Cares Nepal Huo ma cao China Chanvre cultive France Huo ma China Chanvre de l’Inde France Indian hemp United Kingdom Chanvre France Indische hennep Netherlands Chanvrier sauvage France Indischer hanf Germany Charas India Indisk hamp Sweden Churras India Kannabis Finland Da ma cao China Kannabisu Japan Da ma ren China Kerp Albania Da ma China Kinnab Turkey Dagga South Africa Konopie siewne Poland Dansk pot Denmark Konopie Poland Echter hanf Germany Konoplja Slovenia Esrar Turkey Kultur hanf Germany Gaanjaa Nepal Maconha Portugal Gajiimaa Nepal Marihana Netherlands Ganja Guyana Marihouava Greece Ganja India Marihuana Poland Grifa Spain Marihuana Bulgaria Hachis Spain Marihuana Croatia Hamp Denmark Marihuana Czech Republic Hamp Norway Marihuana Denmark Hampa Sweden Marihuana France Hampjurt Iceland Marihuana Germany Hamppu Finland Marihuana Hungary Hanf Germany Marihuana Mexico From: Medicinal Plants of the World, vol. 3: Chemical Constituents, Traditional and Modern Medicinal Uses By: I. A. Ross © Humana Press Inc., Totowa, NJ 29 30 MEDICINAL PLANTS OF THE WORLD Marihuana Russia Porkanchaa Thailand Marihuana Serbia Pot Denmark Marihuana Spain Qinnib Arabic countries Marihuana Ukraine Riesen hanf Germany Marihuana United States Seruma erva Portugal Marijuana France Taima Japan Marijuana Italy Til Arabic countries Marijuana Mexico Vrai chanvre France Marijuana Portugal Weed Guyana Marijuana Sweden Xian ma China Mashinin Japan Ye ma China Navadna konoplja Slovenia BOTANICAL DESCRIPTION types of plants are numerous varieties that Cannabis sativa is an annual herb of the differ from the main one in height, extent MORACEAE family that grows to 5 m tall. -

Altered Motor Development Following Late Gestational Alcohol and Cannabinoid
bioRxiv preprint doi: https://doi.org/10.1101/513713; this version posted January 7, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Altered Motor Development Following Late Gestational Alcohol and Cannabinoid Exposure in Rats Kristen R. Breit, Brandonn Zamudio, & Jennifer D. Thomas Department of Psychology, Center for Behavioral Teratology, San Diego State University, San Diego, CA, 92120, USA Please address correspondence to: Kristen R. Breit, Ph.D. Email: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/513713; this version posted January 7, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 2 Abstract Cannabis is the most commonly used illicit drug among pregnant women, and rates are likely to increase given recent legalization. In addition, half of pregnant women who report consuming cannabis also report drinking alcohol. However, little is known about the consequences of prenatal cannabis alone or combination with alcohol, particularly with cannabis products that are continually increasing in potency of the primary psychoactive constituent in cannabis, Δ9-tetrahydrocannabinol (THC). The current study investigated the effects of early exposure to cannabinoids during the brain growth spurt on early physical and motor development alone (Experiment 1) or in combination with alcohol (Experiment 2). In Experiment 1, Sprague-Dawley rat pups were exposed to a cannabinoid receptor agonist (CP-55,940 [CP]; 0.1, 0.25, 0.4 mg/kg/day), the drug vehicle, or a saline control from postnatal days (PD) 4-9. -

Fast Detection of 10 Cannabinoids by RP-HPLC-UV Method in Cannabis Sativa L
molecules Article Fast Detection of 10 Cannabinoids by RP-HPLC-UV Method in Cannabis sativa L. Mara Mandrioli 1, Matilde Tura 1, Stefano Scotti 2 and Tullia Gallina Toschi 1,* 1 Department of Agricultural and Food Sciences, Alma Mater Studiorum-University of Bologna, Viale Fanin 40, 40127 Bologna, Italy; [email protected] (M.M.); [email protected] (M.T.) 2 Shimadzu Italia, Via G. B. Cassinis 7, 20139 Milano, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-051-209-6010 Academic Editor: Marcello Locatelli Received: 10 May 2019; Accepted: 31 May 2019; Published: 4 June 2019 Abstract: Cannabis has regained much attention as a result of updated legislation authorizing many different uses and can be classified on the basis of the content of tetrahydrocannabinol (THC), a psychotropic substance for which there are legal limitations in many countries. For this purpose, accurate qualitative and quantitative determination is essential. The relationship between THC and cannabidiol (CBD) is also significant as the latter substance is endowed with many specific and non-psychoactive proprieties. For these reasons, it becomes increasingly important and urgent to utilize fast, easy, validated, and harmonized procedures for determination of cannabinoids. The procedure described herein allows rapid determination of 10 cannabinoids from the inflorescences of Cannabis sativa L. by extraction with organic solvents. Separation and subsequent detection are by RP-HPLC-UV. Quantification is performed by an external standard method through the construction of calibration curves using pure standard chromatographic reference compounds. The main cannabinoids dosed (g/100 g) in actual samples were cannabidiolic acid (CBDA), CBD, and D9-THC (Sample L11 CBDA 0.88 0.04, CBD 0.48 0.02, D9-THC 0.06 0.00; Sample L5 CBDA ± ± ± 0.93 0.06, CBD 0.45 0.03, D9-THC 0.06 0.00). -

Invited Review Article การใช้กัญชาเพื่อประโยชน์ทางการแพทย์
Medical Uses of Cannabis IJPS Sripanidkulchai B. Vol. 15 No. 4 October – December 2019 _______________________________________________________________________________________________ Invited Review Article การใช้กัญชาเพื่อประโยชน์ทางการแพทย์ 1 บงั อร ศรีพานิชกลุ ชยั * 1 ศูนย์วิจัยและพัฒนาผลิตภัณฑ์สุขภาพจากสมุนไพร คณะเภสัชศาสตร์ มหาวิทยาลัยขอนแก่น ต.ในเมือง อ.เมือง จ.ขอนแก่น 40002 * ติดต่อผนู้ ิพนธ ์ : ศูนย์วิจัยและพัฒนาผลิตภัณฑ์สุขภาพจากสมุนไพร คณะเภสัชศาสตร์ มหาวิทยาลัยขอนแก่น ต.ในเมือง อ.เมือง จ.ขอนแก่น 40002 อีเมล: [email protected] บทคดั ย่อ การใช้กัญชาเพื่อประโยชน์ทางการแพทย์ บังอร ศรีพานิชกุลชัย1* ว. เภสัชศาสตร์อีสาน 2562; 15(4) : 1-26 รับบทความ : 11 กันยายน 2562 แก้ไขบทความ: 21 ตุลาคม 2562 ตอบรับ: 14 พฤศจิกายน 2562 กัญชาเป็นพืชในวงศ์แคนนาบาซีที่ขึ้นได้ดีในเขตอากาศร้อน เช่น ประเทศไทย และจัดเป็นพืชเสพติดที่เคยใช้เป็นยารักษาโรค แต่ต่อมามกี ารหา้ มใชเ้ น่ืองจากทา ใหเ้ กดิ การเสพตดิ การศกึ ษาต่อมาพบว่าสารสา คญั กลุ่มแคนนาบนิ อยดม์ ฤี ทธติ์ ่อร่างกายหลายประการ โดยที่สารสาคัญที่อยู่ในความสนใจคือ ทีเอชซี (delta-9-tetracannabinol, THC) ซง่ึ มฤี ทธติ์ ่อจติ ประสาทและซบี ดี ี (cannabidiol, CBD) ที่ ไม่มฤี ทธติ์ ่อจติ ประสาท ปัจจุบนั มผี ลงานวจิ ยั ช้แี นะว่าสารทงั้ สองชนิดสามารถน ามาใช้เป็นยารกั ษาโรคและอาการแสดงไดห้ ลายชนิด ได้แก่ อาการปวดประสาท อาการหดเกร็งของกล้ามเนื้อ ต้านการชัก อาการคลื่นไส้อาเจียน เพิ่มความอยากอาหาร โดยมีกลไกการทางาน ผ่านตัวรับเอนโดแคนตาบินอยด์ที่มีบทบาทสาคัญในการควบคุมภาวะโฮมิโอสิสของร่างกาย บทความนี้จึงมีวัตถุประสงค์ที่จะรวบรวมสรุป องค์ความรู้ที่เป็นปัจจุบันเกี่ยวกับกัญชาเพื่อให้เกิดความเข้าใจที่ถูกต้องในด้านการน ากัญชามาใช้เป็นประโยชน์ทางการแพทย์ -
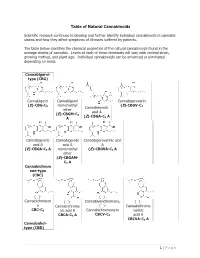
Table of Natural Cannabinoids
Table of Natural Cannabinoids Scientific research continues to develop and further identify individual cannabinoids in cannabis strains and how they affect symptoms of illnesses suffered by patients. The table below identifies the chemical properties of the natural cannabinoids found in the average strains of cannabis. Levels of each of these chemicals will vary with varietal strain, growing method, and plant age. Individual cannabinoids can be enhanced or eliminated depending on need. Cannabigerol- type (CBG) Cannabigerol Cannabigerol Cannabigerovarin (E)-CBG-C monomethyl (E)-CBGV-C 5 Cannabinerolic 3 ether acid A (E)-CBGM-C 5 (Z)-CBGA-C A A 5 Cannabigerolic Cannabigerolic Cannabigerovarinic acid acid A acid A A (E)-CBGA-C5 A monomethyl (E)-CBGVA-C3 A ether (E)-CBGAM- C5 A Cannabichrom ene-type (CBC) (±)- (±)- Cannabichromen (±)- Cannabivarichromene, (±)- e Cannabichrome (±)- Cannabichrome CBC-C5 nic acid A Cannabichromevarin varinic CBCA-C5 A CBCV-C3 acid A CBCVA-C3 A Cannabidiol- type (CBD) 1 | Page (−)-Cannabidiol Cannabidiol Cannabidiol-C4 (−)- Cannabidiorc CBD-C5 momomethyl CBD-C4 Cannabidivarin ol ether CBDV-C3 CBD-C1 CBDM-C5 Cannabidiolic Cannabidivarini acid c acid CBDA-C5 CBDVA-C3 Cannabinodiol- type (CBND) Cannabinodiol Cannabinodivar CBND-C5 in CBND-C3 Tetrahydrocan nabinol-type (THC) 9 9 9 Δ - Δ - Δ - Δ9- Tetrahydrocanna Tetrahydrocan Tetrahydrocannabivarin 9 Tetrahydrocan binol nabinol-C4 Δ -THCV-C3 9 9 nabiorcol Δ -THC-C5 Δ -THC-C4 9 Δ -THCO-C1 9 9 Δ -Tetrahydro- Δ9-Tetrahydro- Δ -Tetrahydro- Δ9-Tetrahydro- cannabinolic