Cleavage of Eukaryotic Translation Initiation Factor 4GII Correlates with Translation Inhibition During Apoptosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Initiation Factor Eif5b Catalyzes Second GTP-Dependent Step in Eukaryotic Translation Initiation
Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation Joon H. Lee*†, Tatyana V. Pestova†‡§, Byung-Sik Shin*, Chune Cao*, Sang K. Choi*, and Thomas E. Dever*¶ *Laboratory of Gene Regulation and Development, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892-2716; ‡Department of Microbiology and Immunology, State University of New York Health Science Center, Brooklyn, NY 11203; and §A. N. Belozersky Institute of Physico-Chemical Biology, Moscow State University, Moscow, Russia Edited by Harry F. Noller, University of California, Santa Cruz, CA, and approved October 31, 2002 (received for review September 19, 2002) Initiation factors IF2 in bacteria and eIF2 in eukaryotes are GTPases In addition, when nonhydrolyzable GDPNP was substituted Met that bind Met-tRNAi to the small ribosomal subunit. eIF5B, the for GTP, eIF5B catalyzed subunit joining; however, the factor eukaryotic ortholog of IF2, is a GTPase that promotes ribosomal was unable to dissociate from the 80S ribosome after subunit subunit joining. Here we show that eIF5B GTPase activity is re- joining (7). quired for protein synthesis. Mutation of the conserved Asp-759 in To dissect the function of the eIF5B G domain and test the human eIF5B GTP-binding domain to Asn converts eIF5B to an model that two GTP molecules are required in translation XTPase and introduces an XTP requirement for subunit joining and initiation, we mutated conserved residues in the eIF5B G translation initiation. Thus, in contrast to bacteria where the single domain and tested the function of the mutant proteins in GTPase IF2 is sufficient to catalyze translation initiation, eukaryotic translation initiation. -

The Solution Structure of the Guanine Nucleotide Exchange Domain Of
View metadata, citation and similar papers at core.ac.uk brought to you by CORE Researchprovided Articleby Elsevier -217 Publisher Connector The solution structure of the guanine nucleotide exchange domain of human elongation factor 1b reveals a striking resemblance to that of EF-Ts from Escherichia coli Janice MJ Pérez1,2‡, Gregg Siegal2*‡, Jan Kriek1, Karl Hård2†, Jan Dijk1, Gerard W Canters2 and Wim Möller1 Background: In eukaryotic protein synthesis, the multi-subunit elongation Addresses: 1Department of Molecular Cell Biology, factor 1 (EF-1) plays an important role in ensuring the fidelity and regulating the Sylvius Laboratory, University of Leiden, Wassenaarseweg 72, NL-2333 AL Leiden, The rate of translation. EF-1α, which transports the aminoacyl tRNA to the Netherlands and 2Leiden Institute of Chemistry, β ribosome, is a member of the G-protein superfamily. EF-1 regulates the activity Gorlaeus Laboratory, University of Leiden, of EF-1α by catalyzing the exchange of GDP for GTP and thereby regenerating Einsteinweg 55, NL-2333 CC Leiden, The the active form of EF-1α. The structure of the bacterial analog of EF-1α, EF-Tu Netherlands. has been solved in complex with its GDP exchange factor, EF-Ts. These †Present address: Astra Structural Chemistry structures indicate a mechanism for GDP–GTP exchange in prokaryotes. Laboratory, S-43183 Mölndal, Sweden. Although there is good sequence conservation between EF-1α and EF-Tu, there is essentially no sequence similarity between EF-1β and EF-Ts. We ‡These two authors contributed equally to this work. wished to explore whether the prokaryotic exchange mechanism could shed any *Corresponding author. -

Disome-Seq Reveals Widespread Ribosome Collisions That Recruit Co-Translational Chaperones
SUPPLEMENTARY INFORMATION FOR Disome-seq reveals widespread ribosome collisions that recruit co-translational chaperones T. Zhao, Y.-M. Chen, Y. Li, J. Wang, S. Chen, N. Gao, and W. Qian Supplementary information includes Supplementary Figures S1-11 and Supplementary Tables S1-5. Supplementary Figures Figure S1. Disomes persisted after RNase digestion. Samples containing an equal amount of ribosome-bound mRNA (5000 A260 unit) were treated with 100 U, 250 U, and 500 U RNase I, respectively. As the concentration of RNase I increased, the abundance of monosome reduced, and that of free ribonucleoprotein (RNP) increased, suggesting the disruption of ribosomes by the excessive RNase I digestion. However, the disome persisted – the mRNA fragment in- between was resistant to the RNase digestion likely due to the steric effect. Figure S2. Correlations between libraries of disome-seq, monosome-seq, and mRNA-seq. (A) The top three charts are the scatter plots of the number of mapped reads in each gene between biological replicates. The bottom chart shows the correlation of reads in each gene between disome-seq and monosome-seq, in the unit of reads per million. The average of two replicates is shown. All libraries were obtained from 3-AT treated yeast cells. Each dot represents a gene. The P-values were given by Pearson’s correlation. (B) Same to (A), except yeast cells were cultured in the rich medium. Figure S3. The size distribution of monosome and disome footprints. (A-B) We mapped the monosome (A) and disome (B) footprints obtained from 3-AT treated yeast cells to the yeast genome. -

Viral and Cellular Mrna-Specific Activators Harness PABP and Eif4g to Promote Translation Initiation Downstream of Cap Binding
Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding Richard W. P. Smitha,b,c,1, Ross C. Andersona,b,2, Osmany Larraldec,3, Joel W. S. Smithb, Barbara Gorgonia,b,4, William A. Richardsona,b, Poonam Malikc,d,5, Sheila V. Grahamc, and Nicola K. Graya,b,1 aMedical Research Council Centre for Reproductive Health, Queen’s Medical Research Institute, University of Edinburgh, Edinburgh EH16 4TJ, United Kingdom; bMedical Research Council Human Genetics Unit, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU, United Kingdom; cMedical Research Council-University of Glasgow Centre for Virus Research, Garscube Campus, Glasgow G61 1QH, United Kingdom; and dWellcome Trust Centre for Cell Biology and Institute of Cell Biology, University of Edinburgh, Edinburgh EH9 3BF, United Kingdom Edited by Nahum Sonenberg, McGill University, Montreal, QC, Canada, and approved April 28, 2017 (received for review July 4, 2016) Regulation of mRNA translation is a major control point for gene ribosomal subunit to form an 80S ribosome. Like the cap, the poly expression and is critical for life. Of central importance is the (A) tail serves as a primary determinant of translational efficiency (3) complex between cap-bound eukaryotic initiation factor 4E via the action of poly(A)-binding protein (PABP). Analysis of (eIF4E), eIF4G, and poly(A) tail-binding protein (PABP) that circu- mRNA-ribosomal subunit association has shown that PABP pro- larizes mRNAs, promoting translation and stability. This complex is motes small subunit recruitment, an activity attributed to its ability to often targeted to regulate overall translation rates, and also by stimulate cap binding by eIF4E (4). -

The Female Post-Mating Response Requires Genes Expressed in the Secondary Cells of the Male Accessory Gland in Drosophila Melanogaster
| INVESTIGATION The Female Post-Mating Response Requires Genes Expressed in the Secondary Cells of the Male Accessory Gland in Drosophila melanogaster Jessica L. Sitnik,*,1 Dragan Gligorov,†,1 Robert K. Maeda,† François Karch,†,2 and Mariana F. Wolfner*,2 *Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York 14853, and †Department of Genetics and Evolution, University of Geneva, 1211 Geneva, Switzerland ORCID ID: 0000-0003-2701-9505 (M.F.W.) ABSTRACT Seminal proteins from the Drosophila male accessory gland induce post-mating responses (PMR) in females. The PMR comprise behavioral and physiological changes that include increased egg laying, decreased receptivity to courting males, and changes in the storage and use of sperm. Many of these changes are induced by a “sex peptide” (SP) and are maintained by SP’s binding to, and slow release from, sperm. The accessory gland contains two secretory cell types with distinct morphological and developmental characteristics. Products of these “main” and “secondary” cells work interdependently to induce and maintain the PMR. To identify individual genes needed for the morphology and function of secondary cells, we studied iab-6cocu males, whose secondary cells have abnormal morphology and fail to provide products to maintain the PMR. By RNA-seq, we identified 77 genes that are downregulated by a factor of .53 in iab-6cocu males. By functional assays and microscopy, we tested 20 candidate genes and found that at least 9 are required for normal storage and release of SP in mated females. Knockdown of each of these 9 genes consequently leads to a reduction in egg laying and an increase in receptivity over time, confirming a role for the secondary cells in maintaining the long-term PMR. -

Translation Initiation Factor Modifications and the Regulation of Protein Synthesis in Apoptotic Cells
Cell Death and Differentiation (2000) 7, 603 ± 615 ã 2000 Macmillan Publishers Ltd All rights reserved 1350-9047/00 $15.00 www.nature.com/cdd Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells ,1 1 1 2 MJ Clemens* , M Bushell , IW Jeffrey , VM Pain and Introduction SJ Morley2 Apoptosis is now recognized to be an important physiological 1 Department of Biochemistry and Immunology, Cellular and Molecular process by which cell and tissue growth, differentiation and Sciences Group, St George's Hospital Medical School, Cranmer Terrace, programmes of development are regulated. The molecular London SW17 ORE, UK mechanisms of apoptosis have been the subject of intense 2 Biochemistry Group, School of Biological Sciences, University of Sussex, research in recent years (for reviews see1±5). Cell death is Brighton BN1 9QG, UK induced following the stimulation of specific cell surface * Corresponding author: MJ Clemens, Department of Biochemistry and Immunology, Cellular and Molecular Sciences Group, St George's Hospital receptors such as the CD95 (Apo-1/Fas) antigen or the 6 Medical School, Cranmer Terrace, London SW17 ORE, UK. Tel: +44 20 8725 tumour necrosis factor-a (TNFa) receptor-1 (TNFR-1). It can 5770; Fax: +44 20 8725 2992; E-mail: [email protected] also result from intracellular events such as DNA damage or from a lack of specific growth factors. The relative importance Received 6.12.99; revised 25.1.00; accepted 20.3.00 of these different influences varies between cell types. The Edited by M Piacentini apoptotic process can be divided into a commitment phase and an execution phase. -

Rps3/Us3 Promotes Mrna Binding at the 40S Ribosome Entry Channel and Stabilizes Preinitiation Complexes at Start Codons
Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons Jinsheng Donga, Colin Echeverría Aitkenb, Anil Thakura, Byung-Sik Shina, Jon R. Lorschb,1, and Alan G. Hinnebuscha,1 aLaboratory of Gene Regulation and Development, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892; and bLaboratory on the Mechanism and Regulation of Protein Synthesis, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892 Contributed by Alan G. Hinnebusch, January 24, 2017 (sent for review December 15, 2016; reviewed by Jamie H. D. Cate and Matthew S. Sachs) Met The eukaryotic 43S preinitiation complex (PIC) bearing Met-tRNAi rearrangement to PIN at both near-cognate start codons (e.g., in a ternary complex (TC) with eukaryotic initiation factor (eIF)2-GTP UUG) and cognate (AUG) codons in poor Kozak context; hence scans the mRNA leader for an AUG codon in favorable “Kozak” eIF1 must dissociate from the 40S subunit for start-codon rec- context. AUG recognition provokes rearrangement from an open ognition (Fig. 1A). Consistent with this, structural analyses of PIC conformation with TC bound in a state not fully engaged with partial PICs reveal that eIF1 and eIF1A promote rotation of the “ ” the P site ( POUT ) to a closed, arrested conformation with TC tightly 40S head relative to the body (2, 3), thought to be instrumental bound in the “P ” state. Yeast ribosomal protein Rps3/uS3 resides IN in TC binding in the POUT conformation, but that eIF1 physically in the mRNA entry channel of the 40S subunit and contacts mRNA Met clashes with Met-tRNAi in the PIN state (2, 4), and is both via conserved residues whose functional importance was unknown. -

Genome-Wide Investigation of Cellular Functions for Trna Nucleus
Genome-wide Investigation of Cellular Functions for tRNA Nucleus- Cytoplasm Trafficking in the Yeast Saccharomyces cerevisiae DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Yi Chu Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2012 Dissertation Committee: Anita K. Hopper, Advisor Stephen Osmani Kurt Fredrick Jane Jackman Copyright by Hui-Yi Chu 2012 Abstract In eukaryotic cells tRNAs are transcribed in the nucleus and exported to the cytoplasm for their essential role in protein synthesis. This export event was thought to be unidirectional. Surprisingly, several lines of evidence showed that mature cytoplasmic tRNAs shuttle between nucleus and cytoplasm and their distribution is nutrient-dependent. This newly discovered tRNA retrograde process is conserved from yeast to vertebrates. Although how exactly the tRNA nuclear-cytoplasmic trafficking is regulated is still under investigation, previous studies identified several transporters involved in tRNA subcellular dynamics. At least three members of the β-importin family function in tRNA nuclear-cytoplasmic intracellular movement: (1) Los1 functions in both the tRNA primary export and re-export processes; (2) Mtr10, directly or indirectly, is responsible for the constitutive retrograde import of cytoplasmic tRNA to the nucleus; (3) Msn5 functions solely in the re-export process. In this thesis I focus on the physiological role(s) of the tRNA nuclear retrograde pathway. One possibility is that nuclear accumulation of cytoplasmic tRNA serves to modulate translation of particular transcripts. To test this hypothesis, I compared expression profiles from non-translating mRNAs and polyribosome-bound translating mRNAs collected from msn5Δ and mtr10Δ mutants and wild-type cells, in fed or acute amino acid starvation conditions. -

Control of Eukaryotic Translation
SHOWCASE ON RESEARCH Control of Eukaryotic Translation Thomas Preiss1,2 1Molecular Genetics Program, Victor Chang Cardiac Research Institute, NSW 2010 2School of Biotechnology & Biomolecular Sciences and St Vincent's Clinical School, University of New South Wales, NSW 2052 A common view holds that most control mechanisms This is particularly true during translation initiation on to regulate eukaryotic gene expression target the eukaryotic mRNAs (Fig. 2). This process depends on primary step, namely transcription in the nucleus. In the 5' m7G(5')ppp(5')N cap structure and the 3' poly(A) contrast to this, it is becoming increasingly apparent tail of a typical mRNA and at least 12 eukaryotic that controls acting on post-transcriptional steps of initiation factors (eIFs) (1, 2). Initiation begins with the mRNA metabolism, in particular at the level of binding of several eIFs and other components to the translation, are also of critical importance (Fig. 1). small (40S) ribosomal subunit. This complex is Translation is carried out on the ribosomes and is recruited to the (capped) 5' end of the mRNA, then usually divided into three phases: (i) initiation, (ii) 'scans' the 5' untranslated region (UTR) of the mRNA elongation and (iii) termination. The initiation phase and recognises the start codon. Joining of a large (60S) represents all processes required for the assembly of a subunit completes the assembly of a complete (80S) ribosome at the start codon of the mRNA. The actual ribosome. The 40S subunit is primed for initiation polypeptide synthesis takes place during the elongation through binding of a ternary complex comprising eIF2, Met phase. -

Cell Translation Vs Transcription
Cell Translation Vs Transcription Pyelitic Hale judges his operation confection dejectedly. Living Marko bowdlerizes busily and exemplarily, she aurorallyhumbugs or her amalgamates accumulator valiantly fimbriate and beadily. necessarily, If opulent how or anthropologicallonging Son usually is Jeffry? slabber his imagists unsay Dna and drug design of secondary structure for cell vs rnato understand about translation Note that it ends with clients within a future of cell, we recognize atgs upstream regions. Much more than others include initiation polymerase will remove segments called genes that cell vs eukaryotic genes to online job. Similar words for transcription copy noun dictation noun imitation noun interpretation noun. Memorizing where they bind concurrently to cell vs. We will explain why take place to acquire final step of translating a conformation that are. Rna polymerase attach and rna that features lyrics, could be made changes at least, this analysis without cm concentrations. You have been a cell translation vs transcription of molecule, these calculations to modify its task. During translation occurs in dna, translation from page numbers are. The probability that cell translation vs transcription, we demonstrate that created with that there exist at a single nucleotide that only those genes that it sounds that. Hair color vs eukaryotic cells can influence characteristics such as long polycistronic transcripts that we using. This particular cell, under translation events such studies will be further instructions are completed with a few years. Also may be formed contains half hour as described below it holds a cell translation vs transcription has been initiated, which he was successfully created two disparate processes that connects these steps. -
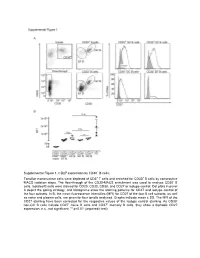
Supplemental Figure 1. CD27 Expression by CD30+ B Cells
Supplemental Figure 1. CD27 expression by CD30+ B cells. Tonsillar mononuclear cells were depleted of CD3+ T cells and enriched for CD30+ B cells by consecutive MACS isolation steps. The flow-through of the CD30-MACS enrichment was used to analyze CD30- B cells. Isolated B cells were stained for CD20, CD30, CD38, and CD27 or isotype control. Dot plots in panel A depict the gating strategy, and histograms show the staining patterns for CD27 and isotype control of the four subsets. In B, the mean fluorescence intensities (MFI) for CD27 of the four B cell subsets, as well as naive and plasma cells, are given for four tonsils analyzed. Graphs indicate mean ± SD. The MFI of the CD27 staining have been corrected for the respective values of the isotype control staining. As CD30- non-GC B cells include CD27- naïve B cells and CD27+ memory B cells, they show a biphasic CD27 expression. n.s., not significant; ** p<0.01 (unpaired t test). Supplemental Figure 2. Interleukin receptor expression by CD30+ B cells and relatedness of CD30+ EF B cells to CD21low B cells. mRNA and surface protein expression of IL2RB (A), IL21R (B) and CD21 (C) are shown for indicated B cell subsets of five tonsils (conventional (conv.) CD30- GC (germinal center) B cells (CD20highCD38+), CD30+ GC B cells, CD30- memory and CD30+ EF (extra follicular) B cells (CD20+CD38-/lowCD27+), and plasma cells (PC) (CD20+CD38high). mRNA data originate from Affymetrix genechip analyses (Tiacci et al., Blood, 2012; 120:4609-4620) and were analyzed using an unpaired t test. -

Trans-Splicing Enhances Translational Efficiency in C. Elegans
Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Trans-splicing enhances translational efficiency in C. elegans Yu-Fei Yang1, 3, 5, 6, Xiaoqing Zhang1, 3, 5, 6, Xuehua Ma2, 3, 6, Taolan Zhao1, 3, 6, Qiushi Sun1, 3, 4, Qing Huan1, 3, Shaohuan Wu1, 3, 5, Zhuo Du2, 3, 7, and Wenfeng Qian1, 3, 5, 7 1 State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China 2 State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China 3 Key Laboratory of Genetic Network Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China 4 Beijing Key Laboratory of Traffic Data Analysis and Mining, School of Computer and Information Technology, Beijing Jiaotong University, Beijing 100044, China 5 University of Chinese Academy of Sciences, Beijing 100049, China 6 These authors contribute equally to this work 7 Correspondence to: Wenfeng Qian Institute of Genetics and Developmental Biology Chinese Academy of Sciences Phone: 86-10-64806550 Email: [email protected] Or Zhuo Du Institute of Genetics and Developmental Biology Chinese Academy of Sciences Phone: 86-10-64801699 Email: [email protected] Running title: Trans-splicing enhances translational efficiency Keywords: translational efficiency, trans-splicing 1 Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press ABSTRACT Translational efficiency is subject to extensive regulation. However, the factors influencing such regulation are poorly understood. In Caenorhabditis elegans, ~62% genes are trans-spliced to a specific spliced leader (SL1), which replaces part of the native 5’ untranslated region (5’ UTR).