Phylogenetic Relationship and Domain Organisation of SET Domain Proteins of Archaeplastida Supriya Sarma* and Mukesh Lodha*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gymnosperms the MESOZOIC: ERA of GYMNOSPERM DOMINANCE
Chapter 24 Gymnosperms THE MESOZOIC: ERA OF GYMNOSPERM DOMINANCE THE VASCULAR SYSTEM OF GYMNOSPERMS CYCADS GINKGO CONIFERS Pinaceae Include the Pines, Firs, and Spruces Cupressaceae Include the Junipers, Cypresses, and Redwoods Taxaceae Include the Yews, but Plum Yews Belong to Cephalotaxaceae Podocarpaceae and Araucariaceae Are Largely Southern Hemisphere Conifers THE LIFE CYCLE OF PINUS, A REPRESENTATIVE GYMNOSPERM Pollen and Ovules Are Produced in Different Kinds of Structures Pollination Replaces the Need for Free Water Fertilization Leads to Seed Formation GNETOPHYTES GYMNOSPERMS: SEEDS, POLLEN, AND WOOD THE ECOLOGICAL AND ECONOMIC IMPORTANCE OF GYMNOSPERMS The Origin of Seeds, Pollen, and Wood Seeds and Pollen Are Key Reproductive SUMMARY Innovations for Life on Land Seed Plants Have Distinctive Vegetative PLANTS, PEOPLE, AND THE Features ENVIRONMENT: The California Coast Relationships among Gymnosperms Redwood Forest 1 KEY CONCEPTS 1. The evolution of seeds, pollen, and wood freed plants from the need for water during reproduction, allowed for more effective dispersal of sperm, increased parental investment in the next generation and allowed for greater size and strength. 2. Seed plants originated in the Devonian period from a group called the progymnosperms, which possessed wood and heterospory, but reproduced by releasing spores. Currently, five lineages of seed plants survive--the flowering plants plus four groups of gymnosperms: cycads, Ginkgo, conifers, and gnetophytes. Conifers are the best known and most economically important group, including pines, firs, spruces, hemlocks, redwoods, cedars, cypress, yews, and several Southern Hemisphere genera. 3. The pine life cycle is heterosporous. Pollen strobili are small and seasonal. Each sporophyll has two microsporangia, in which microspores are formed and divide into immature male gametophytes while still retained in the microsporangia. -

Origin of Gibberellin-Dependent Transcriptional Regulation by Molecular Exploitation of a Transactivation Domain in DELLA Proteins
bioRxiv preprint doi: https://doi.org/10.1101/398883; this version posted December 10, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Article - Discoveries Origin of gibberellin-dependent transcriptional regulation by molecular exploitation of a transactivation domain in DELLA proteins Jorge Hernández-García1, Asier Briones-Moreno1, Renaud Dumas2, and Miguel A. Blázquez1 1Instituto de Biología Molecular y Celular de Plantas (IBMCP), CSIC-Universidad Politécnica de Valencia, Campus UPV CPI 8E, Valencia, Spain 2Laboratoire de Physiologie Cellulaire et Végétale, Université Grenoble Alpes, CNRS, Commissariat à l'Energie Atomique et aux Energies Alternatives/Biosciences and Biotechnology Institute of Grenoble, Institut National de la Recherche Agronomique (INRA), Grenoble, France Corresponding author: Miguel A. Blázquez ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/398883; this version posted December 10, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract DELLA proteins are land-plant specific transcriptional regulators known to interact through their C-terminal GRAS domain with over 150 transcription factors in Arabidopsis thaliana. Besides, DELLAs from vascular plants can interact through the N-terminal domain with the gibberellin receptor encoded by GID1, through which gibberellins promote DELLA degradation. However, this regulation is absent in non-vascular land plants, which lack active gibberellins or a proper GID1 receptor. -

General Botany Lab Review Fungi, Algae, Bryophytes, Ferns & Fern Allies
General Botany Lab Review Fungi, Algae, Bryophytes, Ferns & Fern Allies You have looked at a lot of stuff – both live and via prepared slides. You’ve also labeled at least one Life Cycle Diagram for each of the groups. Know what your benchmarks are for a general life cycle diagram and be able to label them. I will not ask you to identify anything to species or genus; be able to identify things to “group” (i.e., ascomycete, bryophyta, etc.) Be able to identify growth form (e.g., unicell, filamentous, etc.). Recognize the differences between sexual and asexual reroductive structures. All questions will be multiple choice. Material looked at: UNIT 1: FUNGI EXERCISE 1: CHYTRIDS/ CHYTRIDOMYCOTA: Allmyces arbusculus – life and prepared slides EXERCISE 2: ZYGOMYCETES/ ZYGOMYCOTA: Rhizopus stolonifer – live and prepared slides EXERCISE 2: MYCORRHIZA and the GLOMEROMYCETES/ GLOMEROMYCOTA – prepared slides only EXERCISE 3: ASCOMYCETES/ ASCOMYCOTA Aspergillus sp., Penicillium sp., Saccharomyces cerevisiae, Peziza sp., Sordaria fimicola, and Morchella sp. – a mixture of live and prepared materials EXERCISE 4: BASIDIOMYCETES/BASIDIOMYCETES Agaricus, Coprinus, Cronartium (a rust), Ustilago (a smut) – slides, fresh, and dried EXERCISE 5: SLIME MOLDS – live and prepared Physarum EXERCISE 6: LICHENS – live and prepared slides be able to identify the various growth forms UNIT 2: ALGAE EXERCISE 1: CYANOBACTERIA Anabaena sp., Nostoc, and Oscillaroria – live and prepared material EXERCISE 2: SUPERGROUP EXCAVATA (Phylum Euglenophyta) – live and prepared material -

XI MOBILE NO:9340839715 CHAPTER-3 Plant Kingd
NAME OF THE TEACHER: SR. RENCY GEORGE SUBJECT: BIOLOGY TOPIC: CHAPTER -1 CLASS: XI MOBILE NO:9340839715 CHAPTER-3 Plant Kingdom Objectives :- Observe plants closely to notice features and characteristics of growth and development. Observe and identify differences in plants and animals. Observe similarities among plants (seeds, roots, stems, leaves, flowers, fruit) Learning Strategies:- • Explain Different systems of Classification. • Differentiate between Artificial and Natural System of Classification. • Explain Algae and its significance. • Differentiate between various classes of Algae. • Explain various modes of reproduction in Algae. RESOURCES: i.Text books ii.Learning Materials iii.Lab manual iv.E-Resources,video ,L.C.D etc… CLICK HERE TO PLAY CONTENT RELATED VIDEO ON YOU TUBE. https://youtu.be/SWlVX1gDd98 https://youtu.be/P_MyyxIQzm4 https://youtu.be/KmbFGIiwP4k https://youtu.be/mepU8gStVpg https://youtu.be/fnE01M0YlTc https://youtu.be/xir7xvLi8XE Contents Introduction Plant kingdom includes algae, bryophytes, pteridophytes, gymnosperms and angiosperms. ... Depending on the type of pigment possessed and the type of stored food, algae are classified into three classes, namely Chlorophyceae, Phaeophyceae and Rhodophyceae. Eukaryotic, multicellular, chlorophyll containing and having cell wall, are grouped under the kingdom Plantae. It is popularly known as plant kingdom. • Phylogenetic system of classification based on evolutionary relationship is presently used for classifying plants. • Numerical Taxonomy use computer by assigning code for each character and analyzing the features. • Cytotaxonomy is based on cytological information like chromosome number, structure and behaviour. • Chemotaxonomy uses chemical constituents of plants to resolve the confusion. Algae: These include the simplest plants which possess undifferentiated or thallus like forms, reproductive organs single celled called gametangia. -

The Associations Between Pteridophytes and Arthropods
FERN GAZ. 12(1) 1979 29 THE ASSOCIATIONS BETWEEN PTERIDOPHYTES AND ARTHROPODS URI GERSON The Hebrew University of Jerusalem, Faculty of Agriculture, Rehovot, Israel. ABSTRACT Insects belonging to 12 orders, as well as mites, millipedes, woodlice and tardigrades have been collected from Pterldophyta. Primitive and modern, as well as general and specialist arthropods feed on pteridophytes. Insects and mites may cause slight to severe damage, all plant parts being susceptible. Several arthropods are pests of commercial Pteridophyta, their control being difficult due to the plants' sensitivity to pesticides. Efforts are currently underway to employ insects for the biological control of bracken and water ferns. Although Pteridophyta are believed to be relatively resistant to arthropods, the evidence is inconclusive; pteridophyte phytoecdysones do not appear to inhibit insect feeders. Other secondary compounds of preridophytes, like prunasine, may have a more important role in protecting bracken from herbivores. Several chemicals capable of adversely affecting insects have been extracted from Pteridophyta. The litter of pteridophytes provides a humid habitat for various parasitic arthropods, like the sheep tick. Ants often abound on pteridophytes (especially in the tropics) and may help in protecting these plants while nesting therein. These and other associations are discussed . lt is tenatively suggested that there might be a difference in the spectrum of arthropods attacking ancient as compared to modern Pteridophyta. The Osmundales, which, in contrast to other ancient pteridophytes, contain large amounts of ·phytoecdysones, are more similar to modern Pteridophyta in regard to their arthropod associates. The need for further comparative studies is advocated, with special emphasis on the tropics. -

CH28 PROTISTS.Pptx
9/29/14 Biosc 41 Announcements 9/29 Review: History of Life v Quick review followed by lecture quiz (history & v How long ago is Earth thought to have formed? phylogeny) v What is thought to have been the first genetic material? v Lecture: Protists v Are we tetrapods? v Lab: Protozoa (animal-like protists) v Most atmospheric oxygen comes from photosynthesis v Lab exam 1 is Wed! (does not cover today’s lab) § Since many of the first organisms were photosynthetic (i.e. cyanobacteria), a LOT of excess oxygen accumulated (O2 revolution) § Some organisms adapted to use it (aerobic respiration) Review: History of Life Review: Phylogeny v Which organelles are thought to have originated as v Homology is similarity due to shared ancestry endosymbionts? v Analogy is similarity due to convergent evolution v During what event did fossils resembling modern taxa suddenly appear en masse? v A valid clade is monophyletic, meaning it consists of the ancestor taxon and all its descendants v How many mass extinctions seem to have occurred during v A paraphyletic grouping consists of an ancestral species and Earth’s history? Describe one? some, but not all, of the descendants v When is adaptive radiation likely to occur? v A polyphyletic grouping includes distantly related species but does not include their most recent common ancestor v Maximum parsimony assumes the tree requiring the fewest evolutionary events is most likely Quiz 3 (History and Phylogeny) BIOSC 041 1. How long ago is Earth thought to have formed? 2. Why might many organisms have evolved to use aerobic respiration? PROTISTS! Reference: Chapter 28 3. -
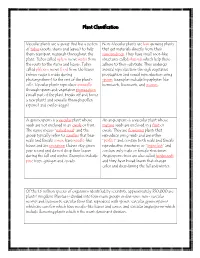
Plant Classification
Plant Classification Vascular plants are a group that has a system Non-Vascular plants are low growing plants of tubes (roots, stems and leaves) to help that get materials directly from their them transport materials throughout the surroundings. They have small root-like plant. Tubes called xylem move water from structures called rhizoids which help them the roots to the stems and leaves. Tubes adhere to their substrate. They undergo called phloem move food from the leaves asexual reproduction through vegetative (where sugar is made during propagation and sexual reproduction using photosynthesis) to the rest of the plant’s spores. Examples include bryophytes like cells. Vascular plants reproduce asexually hornworts, liverworts, and mosses. through spores and vegetative propagation (small part of the plant breaks off and forms a new plant) and sexually through pollen (sperm) and ovules (eggs). A gymnosperm is a vascular plant whose An angiosperm is a vascular plant whose seeds are not enclosed in an ovule or fruit. mature seeds are enclosed in a fruit or The name means “naked seed” and the ovule. They are flowering plants that group typically refers to conifers that bear reproduce using seeds and are either male and female cones, have needle-like “perfect” and contain both male and female leaves and are evergreen (leaves stay green reproductive structures or “imperfect” and year round and do not drop their leaves contain only male or female structures. during the fall and winter. Examples include Angiosperm trees are also called hardwoods pine trees, ginkgos and cycads. and they have broad leaves that change color and drop during the fall and winter. -

B.Sc Botany (Sub.) I Group: a General Characters of Gymnosperm
1 B.Sc Botany (Sub.) I Group: A General Characters of Gymnosperm Gymnosperms are a small group of plants comprising only 70 genera and 725 living species. The word Gymnosperm was used by the Greek botanists Theophrastus in 300 B.C. for the plants with unprotected seeds. Gymnosperms are naked seeded plants. The gymnosperms are characterized by the following features: i. It shows the distinct alternation of generations. ii. Sporophytic generation is the dormant phase of the life cycle. The main plants are sporophyte and the gametophytes are dependent on it throughout. iii. The sporophytic plants are usually tall, woody, perennial trees or shrubs and differentiated into root, stem and leaves. iv. Root- Root is usually well developed tap root system. The stele in root is diarch or polyarch. v. Stem- stems are usually branched but unbranched in Cycas. vi. The leaves may be compound as in Cycas or simple as in Pinus. Internal structure: i. The vascular bundles in stem are arranged in a ring. The bundles are conjoint, collateral and open. ii. The secondary growth is effected by a cambial layer which produces secondary xylem and secondary phloem on the inner and outer side respectively. iii. The secondary wood forming distinct annual ring. Spring wood: It is made up of layer tracheids with the walls and layer lumen. Autumn wood: It is made up of compactly arranged smaller tracheids with thick walls and smaller lumen. iv. The wood may be manoxylic or pycnoxylic and may be monoxylic or polyxylic. Dr. Sanjeev Kumar Vidyarthi, dept. of Botany, Dr. L.K.V.D. -

Pteridophyte Fungal Associations: Current Knowledge and Future Perspectives
This is a repository copy of Pteridophyte fungal associations: Current knowledge and future perspectives. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/109975/ Version: Accepted Version Article: Pressel, S, Bidartondo, MI, Field, KJ orcid.org/0000-0002-5196-2360 et al. (2 more authors) (2016) Pteridophyte fungal associations: Current knowledge and future perspectives. Journal of Systematics and Evolution, 54 (6). pp. 666-678. ISSN 1674-4918 https://doi.org/10.1111/jse.12227 © 2016 Institute of Botany, Chinese Academy of Sciences. This is the peer reviewed version of the following article: Pressel, S., Bidartondo, M. I., Field, K. J., Rimington, W. R. and Duckett, J. G. (2016), Pteridophyte fungal associations: Current knowledge and future perspectives. Jnl of Sytematics Evolution, 54: 666–678., which has been published in final form at https://doi.org/10.1111/jse.12227. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Self-Archiving. Reuse Unless indicated otherwise, fulltext items are protected by copyright with all rights reserved. The copyright exception in section 29 of the Copyright, Designs and Patents Act 1988 allows the making of a single copy solely for the purpose of non-commercial research or private study within the limits of fair dealing. The publisher or other rights-holder may allow further reproduction and re-use of this version - refer to the White Rose Research Online record for this item. Where records identify the publisher as the copyright holder, users can verify any specific terms of use on the publisher’s website. -

Curitiba, Southern Brazil
data Data Descriptor Herbarium of the Pontifical Catholic University of Paraná (HUCP), Curitiba, Southern Brazil Rodrigo A. Kersten 1,*, João A. M. Salesbram 2 and Luiz A. Acra 3 1 Pontifical Catholic University of Paraná, School of Life Sciences, Curitiba 80.215-901, Brazil 2 REFLORA Project, Curitiba, Brazil; [email protected] 3 Pontifical Catholic University of Paraná, School of Life Sciences, Curitiba 80.215-901, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-41-3721-2392 Academic Editor: Martin M. Gossner Received: 22 November 2016; Accepted: 5 February 2017; Published: 10 February 2017 Abstract: The main objective of this paper is to present the herbarium of the Pontifical Catholic University of Parana’s and its collection. The history of the HUCP had its beginning in the middle of the 1970s with the foundation of the Biology Museum that gathered both botanical and zoological specimens. In April 1979 collections were separated and the HUCP was founded with preserved specimens of algae (green, red, and brown), fungi, and embryophytes. As of October 2016, the collection encompasses nearly 25,000 specimens from 4934 species, 1609 genera, and 297 families. Most of the specimens comes from the state of Paraná but there were also specimens from many Brazilian states and other countries, mainly from South America (Chile, Argentina, Uruguay, Paraguay, and Colombia) but also from other parts of the world (Cuba, USA, Spain, Germany, China, and Australia). Our collection includes 42 fungi, 258 gymnosperms, 299 bryophytes, 2809 pteridophytes, 3158 algae, 17,832 angiosperms, and only one type of Mimosa (Mimosa tucumensis Barneby ex Ribas, M. -

23.3 Groups of Protists
Chapter 23 | Protists 639 cysts that are a protective, resting stage. Depending on habitat of the species, the cysts may be particularly resistant to temperature extremes, desiccation, or low pH. This strategy allows certain protists to “wait out” stressors until their environment becomes more favorable for survival or until they are carried (such as by wind, water, or transport on a larger organism) to a different environment, because cysts exhibit virtually no cellular metabolism. Protist life cycles range from simple to extremely elaborate. Certain parasitic protists have complicated life cycles and must infect different host species at different developmental stages to complete their life cycle. Some protists are unicellular in the haploid form and multicellular in the diploid form, a strategy employed by animals. Other protists have multicellular stages in both haploid and diploid forms, a strategy called alternation of generations, analogous to that used by plants. Habitats Nearly all protists exist in some type of aquatic environment, including freshwater and marine environments, damp soil, and even snow. Several protist species are parasites that infect animals or plants. A few protist species live on dead organisms or their wastes, and contribute to their decay. 23.3 | Groups of Protists By the end of this section, you will be able to do the following: • Describe representative protist organisms from each of the six presently recognized supergroups of eukaryotes • Identify the evolutionary relationships of plants, animals, and fungi within the six presently recognized supergroups of eukaryotes • Identify defining features of protists in each of the six supergroups of eukaryotes. In the span of several decades, the Kingdom Protista has been disassembled because sequence analyses have revealed new genetic (and therefore evolutionary) relationships among these eukaryotes. -

Brown Algae and 4) the Oomycetes (Water Molds)
Protista Classification Excavata The kingdom Protista (in the five kingdom system) contains mostly unicellular eukaryotes. This taxonomic grouping is polyphyletic and based only Alveolates on cellular structure and life styles not on any molecular evidence. Using molecular biology and detailed comparison of cell structure, scientists are now beginning to see evolutionary SAR Stramenopila history in the protists. The ongoing changes in the protest phylogeny are rapidly changing with each new piece of evidence. The following classification suggests 4 “supergroups” within the Rhizaria original Protista kingdom and the taxonomy is still being worked out. This lab is looking at one current hypothesis shown on the right. Some of the organisms are grouped together because Archaeplastida of very strong support and others are controversial. It is important to focus on the characteristics of each clade which explains why they are grouped together. This lab will only look at the groups that Amoebozoans were once included in the Protista kingdom and the other groups (higher plants, fungi, and animals) will be Unikonta examined in future labs. Opisthokonts Protista Classification Excavata Starting with the four “Supergroups”, we will divide the rest into different levels called clades. A Clade is defined as a group of Alveolates biological taxa (as species) that includes all descendants of one common ancestor. Too simplify this process, we have included a cladogram we will be using throughout the SAR Stramenopila course. We will divide or expand parts of the cladogram to emphasize evolutionary relationships. For the protists, we will divide Rhizaria the supergroups into smaller clades assigning them artificial numbers (clade1, clade2, clade3) to establish a grouping at a specific level.