Useful Molecular Modelling and Drug Design Softwares and Databases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Latest Stable Copy and Install It With: Chmod+X Chimera- *.Bin&&./Chimera- *.Bin
Tangram Suite Documentation Release 0.0.8 InsiliChem Mar 26, 2019 General instructions 1 What is Tangram Suite 3 2 How to install Tangram Suite5 3 General usage 7 4 FAQ & Known issues 9 5 List of Tans 11 6 Tangram BondOrder 13 7 Tangram QMSetup 15 8 Tangram DummyAtoms 17 9 Tangram GAUDIView 19 10 Tangram MMSetup 21 11 Tangram NCIPlot 23 12 Tangram NormalModes 25 13 Tangram OrbiTraj 27 14 Tangram PLIP 29 15 Tangram PoPMuSiCGUI 31 16 Tangram PropKaGUI 33 17 Tangram ReVina 35 18 Tangram SubAlign 37 19 Tangram TalaDraw 39 i ii Tangram Suite Documentation, Release 0.0.8 It’s composed of several independent graphical interfaces and commands for UCSF Chimera. General instructions 1 Tangram Suite Documentation, Release 0.0.8 2 General instructions CHAPTER 1 What is Tangram Suite In progress 3 Tangram Suite Documentation, Release 0.0.8 4 Chapter 1. What is Tangram Suite CHAPTER 2 How to install Tangram Suite 2.1 Install the full Tangram Suite (recommended) 1 - If you don’t have UCSF Chimera installed, download the latest stable copy and install it with: chmod+x chimera- *.bin&&./chimera- *.bin 2 - Download the latest installer from the releases page (Linux and MacOS only) and run it with: bash tangram-*.sh 2.2 Using the conda meta-package Instead of using the Bash installer, you can use conda (if you are already using it) to create a new environment with the tangram metapackage, which will handle all the dependencies. While in alpha, both insilichem channels are needed (the main one and also the dev label): conda create-n tangram-c insilichem/label/dev-c insilichem-c omnia-c rdkit-c ,!conda-forge tangram 2.3 Updating extensions Each extension will check if there’s a new release available every time you launch it. -

GROMACS: Fast, Flexible, and Free
GROMACS: Fast, Flexible, and Free DAVID VAN DER SPOEL,1 ERIK LINDAHL,2 BERK HESS,3 GERRIT GROENHOF,4 ALAN E. MARK,4 HERMAN J. C. BERENDSEN4 1Department of Cell and Molecular Biology, Uppsala University, Husargatan 3, Box 596, S-75124 Uppsala, Sweden 2Stockholm Bioinformatics Center, SCFAB, Stockholm University, SE-10691 Stockholm, Sweden 3Max-Planck Institut fu¨r Polymerforschung, Ackermannweg 10, D-55128 Mainz, Germany 4Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Nijenborgh 4, NL-9747 AG Groningen, The Netherlands Received 12 February 2005; Accepted 18 March 2005 DOI 10.1002/jcc.20291 Published online in Wiley InterScience (www.interscience.wiley.com). Abstract: This article describes the software suite GROMACS (Groningen MAchine for Chemical Simulation) that was developed at the University of Groningen, The Netherlands, in the early 1990s. The software, written in ANSI C, originates from a parallel hardware project, and is well suited for parallelization on processor clusters. By careful optimization of neighbor searching and of inner loop performance, GROMACS is a very fast program for molecular dynamics simulation. It does not have a force field of its own, but is compatible with GROMOS, OPLS, AMBER, and ENCAD force fields. In addition, it can handle polarizable shell models and flexible constraints. The program is versatile, as force routines can be added by the user, tabulated functions can be specified, and analyses can be easily customized. Nonequilibrium dynamics and free energy determinations are incorporated. Interfaces with popular quantum-chemical packages (MOPAC, GAMES-UK, GAUSSIAN) are provided to perform mixed MM/QM simula- tions. The package includes about 100 utility and analysis programs. -

CESMIX: Center for the Exascale Simulation of Materials in Extreme Environments
CESMIX: Center for the Exascale Simulation of Materials in Extreme Environments Project Overview Youssef Marzouk MIT PSAAP-3 Team 18 August 2020 The CESMIX team • Our team integrates expertise in quantum chemistry, atomistic simulation, materials science; hypersonic flow; validation & uncertainty quantification; numerical algorithms; parallel computing, programming languages, compilers, and software performance engineering 1 Project objectives • Exascale simulation of materials in extreme environments • In particular: ultrahigh temperature ceramics in hypersonic flows – Complex materials, e.g., metal diborides – Extreme aerothermal and chemical loading – Predict materials degradation and damage (oxidation, melting, ablation), capturing the central role of surfaces and interfaces • New predictive simulation paradigms and new CS tools for the exascale 2 Broad relevance • Intense current interest in reentry vehicles and hypersonic flight – A national priority! – Materials technologies are a key limiting factor • Material properties are of cross-cutting importance: – Oxidation rates – Thermo-mechanical properties: thermal expansion, creep, fracture – Melting and ablation – Void formation • New systems being proposed and fabricated (e.g., metallic high-entropy alloys) • May have relevance to materials aging • Yet extreme environments are largely inaccessible in the laboratory – Predictive simulation is an essential path… 3 Demonstration problem: specifics • Aerosurfaces of a hypersonic vehicle… • Hafnium diboride (HfB2) promises necessary temperature -

Molecular Structure Input on the Web Peter Ertl
Ertl Journal of Cheminformatics 2010, 2:1 http://www.jcheminf.com/content/2/1/1 REVIEW Open Access Molecular structure input on the web Peter Ertl Abstract A molecule editor, that is program for input and editing of molecules, is an indispensable part of every cheminfor- matics or molecular processing system. This review focuses on a special type of molecule editors, namely those that are used for molecule structure input on the web. Scientific computing is now moving more and more in the direction of web services and cloud computing, with servers scattered all around the Internet. Thus a web browser has become the universal scientific user interface, and a tool to edit molecules directly within the web browser is essential. The review covers a history of web-based structure input, starting with simple text entry boxes and early molecule editors based on clickable maps, before moving to the current situation dominated by Java applets. One typical example - the popular JME Molecule Editor - will be described in more detail. Modern Ajax server-side molecule editors are also presented. And finally, the possible future direction of web-based molecule editing, based on tech- nologies like JavaScript and Flash, is discussed. Introduction this trend and input of molecular structures directly A program for the input and editing of molecules is an within a web browser is therefore of utmost importance. indispensable part of every cheminformatics or molecu- In this overview a history of entering molecules into lar processing system. Such a program is known as a web applications will be covered, starting from simple molecule editor, molecular editor or structure sketcher. -

Popular GPU-Accelerated Applications
LIFE & MATERIALS SCIENCES GPU-ACCELERATED APPLICATIONS | CATALOG | AUG 12 LIFE & MATERIALS SCIENCES APPLICATIONS CATALOG Application Description Supported Features Expected Multi-GPU Release Status Speed Up* Support Bioinformatics BarraCUDA Sequence mapping software Alignment of short sequencing 6-10x Yes Available now reads Version 0.6.2 CUDASW++ Open source software for Smith-Waterman Parallel search of Smith- 10-50x Yes Available now protein database searches on GPUs Waterman database Version 2.0.8 CUSHAW Parallelized short read aligner Parallel, accurate long read 10x Yes Available now aligner - gapped alignments to Version 1.0.40 large genomes CATALOG GPU-BLAST Local search with fast k-tuple heuristic Protein alignment according to 3-4x Single Only Available now blastp, multi cpu threads Version 2.2.26 GPU-HMMER Parallelized local and global search with Parallel local and global search 60-100x Yes Available now profile Hidden Markov models of Hidden Markov Models Version 2.3.2 mCUDA-MEME Ultrafast scalable motif discovery algorithm Scalable motif discovery 4-10x Yes Available now based on MEME algorithm based on MEME Version 3.0.12 MUMmerGPU A high-throughput DNA sequence alignment Aligns multiple query sequences 3-10x Yes Available now LIFE & MATERIALS& LIFE SCIENCES APPLICATIONS program against reference sequence in Version 2 parallel SeqNFind A GPU Accelerated Sequence Analysis Toolset HW & SW for reference 400x Yes Available now assembly, blast, SW, HMM, de novo assembly UGENE Opensource Smith-Waterman for SSE/CUDA, Fast short -

A Web-Based 3D Molecular Structure Editor and Visualizer Platform
Mohebifar and Sajadi J Cheminform (2015) 7:56 DOI 10.1186/s13321-015-0101-7 SOFTWARE Open Access Chemozart: a web‑based 3D molecular structure editor and visualizer platform Mohamad Mohebifar* and Fatemehsadat Sajadi Abstract Background: Chemozart is a 3D Molecule editor and visualizer built on top of native web components. It offers an easy to access service, user-friendly graphical interface and modular design. It is a client centric web application which communicates with the server via a representational state transfer style web service. Both client-side and server-side application are written in JavaScript. A combination of JavaScript and HTML is used to draw three-dimen- sional structures of molecules. Results: With the help of WebGL, three-dimensional visualization tool is provided. Using CSS3 and HTML5, a user- friendly interface is composed. More than 30 packages are used to compose this application which adds enough flex- ibility to it to be extended. Molecule structures can be drawn on all types of platforms and is compatible with mobile devices. No installation is required in order to use this application and it can be accessed through the internet. This application can be extended on both server-side and client-side by implementing modules in JavaScript. Molecular compounds are drawn on the HTML5 Canvas element using WebGL context. Conclusions: Chemozart is a chemical platform which is powerful, flexible, and easy to access. It provides an online web-based tool used for chemical visualization along with result oriented optimization for cloud based API (applica- tion programming interface). JavaScript libraries which allow creation of web pages containing interactive three- dimensional molecular structures has also been made available. -

Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development
processes Review Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development Outi M. H. Salo-Ahen 1,2,* , Ida Alanko 1,2, Rajendra Bhadane 1,2 , Alexandre M. J. J. Bonvin 3,* , Rodrigo Vargas Honorato 3, Shakhawath Hossain 4 , André H. Juffer 5 , Aleksei Kabedev 4, Maija Lahtela-Kakkonen 6, Anders Støttrup Larsen 7, Eveline Lescrinier 8 , Parthiban Marimuthu 1,2 , Muhammad Usman Mirza 8 , Ghulam Mustafa 9, Ariane Nunes-Alves 10,11,* , Tatu Pantsar 6,12, Atefeh Saadabadi 1,2 , Kalaimathy Singaravelu 13 and Michiel Vanmeert 8 1 Pharmaceutical Sciences Laboratory (Pharmacy), Åbo Akademi University, Tykistökatu 6 A, Biocity, FI-20520 Turku, Finland; ida.alanko@abo.fi (I.A.); rajendra.bhadane@abo.fi (R.B.); parthiban.marimuthu@abo.fi (P.M.); atefeh.saadabadi@abo.fi (A.S.) 2 Structural Bioinformatics Laboratory (Biochemistry), Åbo Akademi University, Tykistökatu 6 A, Biocity, FI-20520 Turku, Finland 3 Faculty of Science-Chemistry, Bijvoet Center for Biomolecular Research, Utrecht University, 3584 CH Utrecht, The Netherlands; [email protected] 4 Swedish Drug Delivery Forum (SDDF), Department of Pharmacy, Uppsala Biomedical Center, Uppsala University, 751 23 Uppsala, Sweden; [email protected] (S.H.); [email protected] (A.K.) 5 Biocenter Oulu & Faculty of Biochemistry and Molecular Medicine, University of Oulu, Aapistie 7 A, FI-90014 Oulu, Finland; andre.juffer@oulu.fi 6 School of Pharmacy, University of Eastern Finland, FI-70210 Kuopio, Finland; maija.lahtela-kakkonen@uef.fi (M.L.-K.); tatu.pantsar@uef.fi -

Quantitative Evaluation of Dissociation Mechanisms in Phenolphthalein and the Related Compounds
J. Comput. Chem. Jpn., Vol. 15, No. 1, pp. 13–21 (2016) ©2016 Society of Computer Chemistry, Japan Technical Paper Quantitative Evaluation of Dissociation Mechanisms in Phenolphthalein and the Related Compounds Toshihiko HANAI Health Research Foundation, Research Institute for Production Development 4F, 15 Shimogamo-morimoto-cho, Sakyo-ku, Kyoto 606-0805, Japan e-mail: [email protected] (Received: October 11, 2015; Accepted for publication: April 14, 2016; Online publication: May 6, 2016) Computational chemistry programs were evaluated as aids to teaching qualitative analytical chemistry. Compu- tational chemical calculations can predict absorption spectra, thus enabling the modeling of indicator dissociation mechanisms by different computational chemical programs using a personal computer. An updated MNDO program among 51 programs was found to be the best predictor to explain the dissociation mechanisms of isobenzofuranones and sulfonephthaleins. Unknown dissociation constants were predicted from atomic partial charges instead of Ham- mett's constants. Keywords: Quantitative analysis of dissociation mechanisms, Isobenzofuranone, Sulfonephthalein, Computational chemistry 1 Introduction the absorption wavelength and electron density changes were not well described. The dissociation mechanisms, maximum How to quantitatively teach qualitative analytical chemistry is wavelengths, and electron density maps of isobenzofuranones a very important subject for analytical chemists. Previously, a and sulfonephthaleins were, therefore, evaluated by in silico method to teach molecular interaction mechanisms in chroma- analysis despite the anticipated poor precision. The experimen- tography was quantitatively achieved using molecular mechan- tally measured dissociation of phenolphthalein is described ics and MOPAC programs [1]. Furthermore, the reaction mech- using four dissociation structures, where the ionization of two anisms of highly sensitive detections were also quantitatively phenolic hydroxyl groups converts the neutral molecular form described [2,3]. -

Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department
Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department Name of FOSS Applications Employability GChemPaint GChemPaint is an editor for 2Dchem- GChemPaint is currently being developed ical structures with a multiple docu- as part of The Chemistry Development ment interface. Kit, and a Standard Widget Tool kit- based GChemPaint application is being developed, as part of Bioclipse. Jmol Jmol applet is used to explore the Jmol is a free, open source molecule viewer structure of molecules. Jmol applet is for students, educators, and researchers used to depict X-ray structures in chemistry and biochemistry. It is cross- platform, running on Windows, Mac OS X, and Linux/Unix systems. For PG Students LaTeX Document markup language and Value addition to academic Skills set. preparation system for Tex typesetting Essential for International paper presentation and scientific journals. For PG student for their project work Scilab Scientific Computation package for Value addition in technical problem numerical computations solving via use of computational methods for engineering problems, Applicable in Chemical, ECE, Electrical, Electronics, Civil, Mechanical, Mathematics etc. For PG student who are taking Physical Chemistry Avogadro Avogadro is a free and open source, Research and Development in Chemistry, advanced molecule editor and Pharmacist and University lecturers. visualizer designed for cross-platform use in computational chemistry, molecular modeling, material science, bioinformatics, etc. Spoken Tutorial Project, IIT Bombay Brochure for Commerce and Commerce IT Name of FOSS Applications / Employability LibreOffice – Writer, Calc, Writing letters, documents, creating spreadsheets, tables, Impress making presentations, desktop publishing LibreOffice – Base, Draw, Managing databases, Drawing, doing simple Mathematical Math operations For Commerce IT Students Drupal Drupal is a free and open source content management system (CMS). -
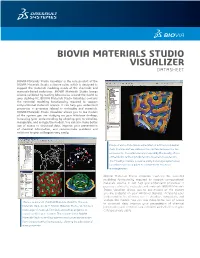
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Report on an NIH Workshop on Ultralarge Chemistry Databases Wendy A
1 Report on an NIH Workshop on Ultralarge Chemistry Databases Wendy A. Warr Wendy Warr & Associates, 6 Berwick Court, Holmes Chapel, Cheshire, CW4 7HZ, United Kingdom. Email: [email protected] Introduction The virtual workshop took place on December 1-3, 2020. It was aimed at researchers, groups, and companies that generate, manage, sell, search, and screen databases of more than one billion small molecules (Figure 1). There were about 550 “attendees” from 37 different countries. Recent advances in computational chemistry have enabled researchers to navigate virtual chemical spaces containing billions of chemical structures, carrying out similarity searches, studying structure-activity relationships (SAR), experimenting with scaffold-hopping, and using other drug discovery methodologies.1 For clarity, one could differentiate “spaces” from “libraries”, and “libraries” from “databases”. Spaces are combinatorially constructed collections of compounds; they are usually very big indeed and it is not possible to enumerate all the precise chemical structures that are covered. Libraries are enumerated collections of full structures: usually fewer than 1010 molecules. Databases are a way to storing libraries, for example, in a relational database management system. Figure 1. Ultralarge chemical databases. (Source: Marcus Gastreich based on the publication by Hoffmann and Gastreich.) This report summarizes talks from about 30 practitioners in the field of ultralarge collections of molecules. The aim is to represent as accurately as possible the information that was delivered by the speakers; the report does not seek to be evaluative. 2 Welcoming remarks; defining a drug discovery gateway Susan Gregurick, Office of Data Science and Strategy, NIH, USA Data should be “findable, accessible, interoperable and reusable” (FAIR)2 and with this in mind, NIH has been creating, curating, integrating, and querying ultralarge chemistry databases. -

Qsar Methods Development, Virtual and Experimental Screening for Cannabinoid Ligand Discovery
QSAR METHODS DEVELOPMENT, VIRTUAL AND EXPERIMENTAL SCREENING FOR CANNABINOID LIGAND DISCOVERY by Kyaw Zeyar Myint BS, Biology, BS, Computer Science, Hampden-Sydney College, 2007 Submitted to the Graduate Faculty of School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2012 UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE This dissertation was presented by Kyaw Zeyar Myint It was defended on August 20th, 2012 and approved by Dr. Ivet Bahar, Professor, Department of Computational and Systems Biology Dr. Billy W. Day, Professor, Department of Pharmaceutical Sciences Dr. Christopher Langmead, Associate Professor, Department of Computer Science, CMU Dissertation Advisor: Dr. Xiang-Qun Xie, Professor, Department of Pharmaceutical Sciences ii Copyright © by Kyaw Zeyar Myint 2012 iii QSAR METHODS DEVELOPMENT, VIRTUAL AND EXPERIMENTAL SCREENING FOR CANNABINOID LIGAND DISCOVERY Kyaw Zeyar Myint, PhD University of Pittsburgh, 2012 G protein coupled receptors (GPCRs) are the largest receptor family in mammalian genomes and are known to regulate wide variety of signals such as ions, hormones and neurotransmitters. It has been estimated that GPCRs represent more than 30% of current drug targets and have attracted many pharmaceutical industries as well as academic groups for potential drug discovery. Cannabinoid (CB) receptors, members of GPCR superfamily, are also involved in the activation of multiple intracellular signal transductions and their endogenous ligands or cannabinoids have attracted pharmacological research because of their potential therapeutic effects. In particular, the cannabinoid subtype-2 (CB2) receptor is known to be involved in immune system signal transductions and its ligands have the potential to be developed as drugs to treat many immune system disorders without potential psychotic side- effects.