A Web-Based 3D Molecular Structure Editor and Visualizer Platform
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Open Data, Open Source, and Open Standards in Chemistry: the Blue Obelisk Five Years On" Journal of Cheminformatics Vol
Oral Roberts University Digital Showcase College of Science and Engineering Faculty College of Science and Engineering Research and Scholarship 10-14-2011 Open Data, Open Source, and Open Standards in Chemistry: The lueB Obelisk five years on Andrew Lang Noel M. O'Boyle Rajarshi Guha National Institutes of Health Egon Willighagen Maastricht University Samuel Adams See next page for additional authors Follow this and additional works at: http://digitalshowcase.oru.edu/cose_pub Part of the Chemistry Commons Recommended Citation Andrew Lang, Noel M O'Boyle, Rajarshi Guha, Egon Willighagen, et al.. "Open Data, Open Source, and Open Standards in Chemistry: The Blue Obelisk five years on" Journal of Cheminformatics Vol. 3 Iss. 37 (2011) Available at: http://works.bepress.com/andrew-sid-lang/ 19/ This Article is brought to you for free and open access by the College of Science and Engineering at Digital Showcase. It has been accepted for inclusion in College of Science and Engineering Faculty Research and Scholarship by an authorized administrator of Digital Showcase. For more information, please contact [email protected]. Authors Andrew Lang, Noel M. O'Boyle, Rajarshi Guha, Egon Willighagen, Samuel Adams, Jonathan Alvarsson, Jean- Claude Bradley, Igor Filippov, Robert M. Hanson, Marcus D. Hanwell, Geoffrey R. Hutchison, Craig A. James, Nina Jeliazkova, Karol M. Langner, David C. Lonie, Daniel M. Lowe, Jerome Pansanel, Dmitry Pavlov, Ola Spjuth, Christoph Steinbeck, Adam L. Tenderholt, Kevin J. Theisen, and Peter Murray-Rust This article is available at Digital Showcase: http://digitalshowcase.oru.edu/cose_pub/34 Oral Roberts University From the SelectedWorks of Andrew Lang October 14, 2011 Open Data, Open Source, and Open Standards in Chemistry: The Blue Obelisk five years on Andrew Lang Noel M O'Boyle Rajarshi Guha, National Institutes of Health Egon Willighagen, Maastricht University Samuel Adams, et al. -

Molecular Structure Input on the Web Peter Ertl
Ertl Journal of Cheminformatics 2010, 2:1 http://www.jcheminf.com/content/2/1/1 REVIEW Open Access Molecular structure input on the web Peter Ertl Abstract A molecule editor, that is program for input and editing of molecules, is an indispensable part of every cheminfor- matics or molecular processing system. This review focuses on a special type of molecule editors, namely those that are used for molecule structure input on the web. Scientific computing is now moving more and more in the direction of web services and cloud computing, with servers scattered all around the Internet. Thus a web browser has become the universal scientific user interface, and a tool to edit molecules directly within the web browser is essential. The review covers a history of web-based structure input, starting with simple text entry boxes and early molecule editors based on clickable maps, before moving to the current situation dominated by Java applets. One typical example - the popular JME Molecule Editor - will be described in more detail. Modern Ajax server-side molecule editors are also presented. And finally, the possible future direction of web-based molecule editing, based on tech- nologies like JavaScript and Flash, is discussed. Introduction this trend and input of molecular structures directly A program for the input and editing of molecules is an within a web browser is therefore of utmost importance. indispensable part of every cheminformatics or molecu- In this overview a history of entering molecules into lar processing system. Such a program is known as a web applications will be covered, starting from simple molecule editor, molecular editor or structure sketcher. -

Getting Started in Jmol
Getting Started in Jmol Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingResources/jmol/jmolTraining/started.html Introduction Physical models of proteins are powerful tools that can be used synergistically with computer visualizations to explore protein structure and function. Although it is interesting to explore models and visualizations created by others, it is much more engaging to create your own! At the MSOE Center for BioMolecular Modeling we use the molecular visualization program Jmol to explore protein and molecular structures in fully interactive 3-dimensional displays. Jmol a free, open source molecular visualization program used by students, educators and researchers internationally. The Jmol Training Guide from the MSOE Center for BioMolecular Modeling will provide the tools needed to create molecular renderings, physical models using 3-D printing technologies, as well as Jmol animations for online tutorials or electronic posters. Examples of Proteins in Jmol Jmol allows users to rotate proteins and molecular structures in a fully interactive 3-dimensional display. Some sample proteins designed with Jmol are shown to the right. Hemoglobin Proteins Insulin Proteins Green Fluorescent safely carry oxygen in the help regulate sugar in Proteins create blood. the bloodstream. bioluminescence in animals like jellyfish. Downloading Jmol Jmol Can be Used in Two Ways: 1. As an independent program on a desktop - Jmol can be downloaded to run on your desktop like any other program. It uses a Java platform and therefore functions equally well in a PC or Mac environment. 2. As a web application - Jmol has a web-based version (oftern refered to as "JSmol") that runs on a JavaScript platform and therefore functions equally well on all HTML5 compatible browsers such as Firefox, Internet Explorer, Safari and Chrome. -

Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department
Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department Name of FOSS Applications Employability GChemPaint GChemPaint is an editor for 2Dchem- GChemPaint is currently being developed ical structures with a multiple docu- as part of The Chemistry Development ment interface. Kit, and a Standard Widget Tool kit- based GChemPaint application is being developed, as part of Bioclipse. Jmol Jmol applet is used to explore the Jmol is a free, open source molecule viewer structure of molecules. Jmol applet is for students, educators, and researchers used to depict X-ray structures in chemistry and biochemistry. It is cross- platform, running on Windows, Mac OS X, and Linux/Unix systems. For PG Students LaTeX Document markup language and Value addition to academic Skills set. preparation system for Tex typesetting Essential for International paper presentation and scientific journals. For PG student for their project work Scilab Scientific Computation package for Value addition in technical problem numerical computations solving via use of computational methods for engineering problems, Applicable in Chemical, ECE, Electrical, Electronics, Civil, Mechanical, Mathematics etc. For PG student who are taking Physical Chemistry Avogadro Avogadro is a free and open source, Research and Development in Chemistry, advanced molecule editor and Pharmacist and University lecturers. visualizer designed for cross-platform use in computational chemistry, molecular modeling, material science, bioinformatics, etc. Spoken Tutorial Project, IIT Bombay Brochure for Commerce and Commerce IT Name of FOSS Applications / Employability LibreOffice – Writer, Calc, Writing letters, documents, creating spreadsheets, tables, Impress making presentations, desktop publishing LibreOffice – Base, Draw, Managing databases, Drawing, doing simple Mathematical Math operations For Commerce IT Students Drupal Drupal is a free and open source content management system (CMS). -

Visualizing 3D Molecular Structures Using an Augmented Reality App
Visualizing 3D molecular structures using an augmented reality app Kristina Eriksen, Bjarne E. Nielsen, Michael Pittelkow 5 Department of Chemistry, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen, Denmark. E-mail: [email protected] ABSTRACT 10 We present a simple procedure to make an augmented reality app to visualize any 3D chemical model. The molecular structure may be based on data from crystallographic data or from computer modelling. This guide is made in such a way, that no programming skills are needed and the procedure uses free software and is a way to visualize 3D structures that are normally difficult to comprehend in the 2D 15 space of paper. The process can be applied to make 3D representation of any 2D object, and we envisage the app to be useful when visualizing simple stereochemical problems, when presenting a complex 3D structure on a poster presentation or even in audio-visual presentations. The method works for all molecules including small molecules, supramolecular structures, MOFs and biomacromolecules. GRAPHICAL ABSTRACT 20 KEYWORDS Augmented reality, Unity, Vuforia, Application, 3D models. 25 Journal 5/18/21 Page 1 of 14 INTRODUCTION Conveying information about three-dimensional (3D) structures in two-dimensional (2D) space, such as on paper or a screen can be difficult. Augmented reality (AR) provides an opportunity to visualize 2D 30 structures in 3D. Software to make simple AR apps is becoming common and ranges of free software now exist to make customized apps. AR has transformed visualization in computer games and films, but the technique is distinctly under-used in (chemical) science.1 In chemical science the challenge of visualizing in 3D exists at several levels ranging from teaching of stereo chemistry problems at freshman university level to visualizing complex molecular structures at 35 the forefront of chemical research. -

Open Data, Open Source and Open Standards in Chemistry: the Blue Obelisk five Years On
Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk ¯ve years on Noel M O'Boyle¤1 , Rajarshi Guha2 , Egon L Willighagen3 , Samuel E Adams4 , Jonathan Alvarsson5 , Richard L Apodaca6 , Jean-Claude Bradley7 , Igor V Filippov8 , Robert M Hanson9 , Marcus D Hanwell10 , Geo®rey R Hutchison11 , Craig A James12 , Nina Jeliazkova13 , Andrew SID Lang14 , Karol M Langner15 , David C Lonie16 , Daniel M Lowe4 , J¶er^omePansanel17 , Dmitry Pavlov18 , Ola Spjuth5 , Christoph Steinbeck19 , Adam L Tenderholt20 , Kevin J Theisen21 , Peter Murray-Rust4 1Analytical and Biological Chemistry Research Facility, Cavanagh Pharmacy Building, University College Cork, College Road, Cork, Co. Cork, Ireland 2NIH Center for Translational Therapeutics, 9800 Medical Center Drive, Rockville, MD 20878, USA 3Division of Molecular Toxicology, Institute of Environmental Medicine, Nobels vaeg 13, Karolinska Institutet, 171 77 Stockholm, Sweden 4Unilever Centre for Molecular Sciences Informatics, Department of Chemistry, University of Cambridge, Lens¯eld Road, CB2 1EW, UK 5Department of Pharmaceutical Biosciences, Uppsala University, Box 591, 751 24 Uppsala, Sweden 6Metamolecular, LLC, 8070 La Jolla Shores Drive #464, La Jolla, CA 92037, USA 7Department of Chemistry, Drexel University, 32nd and Chestnut streets, Philadelphia, PA 19104, USA 8Chemical Biology Laboratory, Basic Research Program, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, USA 9St. Olaf College, 1520 St. Olaf Ave., North¯eld, MN 55057, USA 10Kitware, Inc., 28 Corporate Drive, Clifton Park, NY 12065, USA 11Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260, USA 12eMolecules Inc., 380 Stevens Ave., Solana Beach, California 92075, USA 13Ideaconsult Ltd., 4.A.Kanchev str., So¯a 1000, Bulgaria 14Department of Engineering, Computer Science, Physics, and Mathematics, Oral Roberts University, 7777 S. -

Water and Salt at the Lipid-Solvent Interface
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School April 2019 Water and Salt at the Lipid-Solvent Interface James M. Kruczek University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Physics Commons Scholar Commons Citation Kruczek, James M., "Water and Salt at the Lipid-Solvent Interface" (2019). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/8380 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Water and Salt at the Lipid-Solvent Interface by James M. Kruczek A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Applied Physics Department of Physics College of Arts and Sciences University of South Florida Major Professor: Sagar A. Pandit, Ph.D. Ullah, Ghanim, Ph.D. Robert S. Hoy, Ph.D. Jianjun Pan, Ph.D. Yicheng Tu, Ph.D. Date of Approval: March 26, 2019 Keywords: Lipid Bilayer, Ionic Solvents, Ether Lipids, Molecular Simulations Copyright ⃝c 2018, James M. Kruczek Dedication To my wife Nicole, without whom none of this would be possible. To my father Michael, who labored for his family till his last days. To my family, for all of their support. Acknowledgments The work presented in this document would not be possible without the assistance of many academic professionals. In particular, I would like to acknowledge my major advisor Dr. -

Tracking of Bacterial Metabolism with Azidated Precursors and Click- Chemistry”
Tracking of bacterial metabolism with azidated precursors and click-chemistry Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften vorgelegt beim Fachbereich für Biowissenschaften (15) der Johann Wolfang Goethe Universität in Frankfurt am Main von Alexander J. Pérez aus Nürnberg Frankfurt am Main 2015 Dekanin: Prof. Dr. Meike Piepenbring Gutachter: Prof. Dr. Helge B. Bode Zweitgutachter: Prof. Dr. Joachim W. Engels Datum der Disputation: 2 Danksagung Ich danke meinen Eltern für die stete und vielseitige Unterstützung, deren Umfang ich sehr zu schätzen weiß. Herrn Professor Dr. Helge B. Bode gilt mein besonderer Dank für die Übernahme als Doktorand und für die Gelegenheit meinen Horizont in diesen mich stets faszinierenden Themenbereich in dieser Tiefe erweitern zu lassen. Seine persönliche und fachliche Unterstützung bei der Projektwahl und der entsprechenden Umsetzung ist in dieser Form eine Seltenheit und ich bin mir dieser Tatsache voll bewusst. Gerade die zusätzlich erworbenen Kenntnisse im Bereich der Biologie, sowie der Wert interdisziplinärer Zusammenarbeit ist mir durch zahlreiche freundliche und wertvolle Mitglieder der Arbeitsgruppe bewusst geworden und viele zündenden Ideen wären ohne sie womöglich nie aufgekommen. Einen besonderen Dank möchte ich in diesem Kontext Wolfram Lorenzen und Sebastian Fuchs, die gerade in der Anfangszeit eine große Hilfe waren, ausdrücken. Dies gilt ebenso für die „N100-Crew“ und sämtliche Freunde, die in dieser Zeit zu mir standen und diesen Lebensabschnitt unvergesslich gemacht haben. -

Integrated Multi-Selection Schemes for Complex Molecular Structures
Workshop on Molecular Graphics and Visual Analysis of Molecular Data (MolVA) (2019) J. Byška, M. Krone, and B. Sommer (Editors) MolFind – Integrated Multi-Selection Schemes for Complex Molecular Structures Robin Skånberg1;2, Mathieu Linares1;2, Martin Falk1;2, Ingrid Hotz1;2, and Anders Ynnerman1;2 1Scientific Visualization Group, Linköpings University, Sweden 2Swedish e-Science Research Centre (SeRC) Figure 1: Growing an initial selection of two residues along covalent bonds in a protein fibril. Abstract Selecting components and observing changes of properties and configurations over time is an important step in the analysis of molecular dynamics (MD) data. In this paper, we present a selection tool combining text-based queries with spatial selection and filtering. Morphological operations facilitate refinement of the selection by growth operators, e.g. across covalent bonds. The combination of different selection paradigms enables flexible and intuitive analysis on different levels of detail and visual depiction of molecular events. Immediate visual feedback during interactions ensures a smooth exploration of the data. We demonstrate the utility of our selection framework by analyzing temporal changes in the secondary structure of poly-alanine and the binding of aspirin to phospholipase A2. 1 Introduction In this paper, we present a selection framework for molecu- lar substructures embedded in the visual analytics tool VIA-MD “It is through the interactive manipulation of a visual in- [SLK∗18, KSH∗18]. The work is mostly targeted toward expert terface – the analytic discourse – that knowledge is con- users from the molecular simulation domain. The user-driven de- structed, tested, refined and shared.”[PSCO09] sign of the framework combines a large set of selection paradigms This is also true for the analysis of molecular dynamics (MD) in a flexible way while maintaining a simple and intuitive interface data and the generation of expressive visualization. -
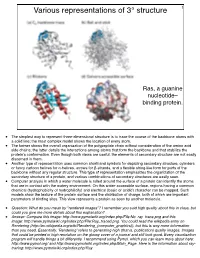
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png. -

Visualizing Protein Structures-Tools and Trends
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 12 January 2020 Visualizing protein structures - tools and trends 1,2 3 1,2 X. Martinez , M. Chavent , M. Baaden 1) CNRS, Université de Paris, UPR 9080, Laboratoire de Biochimie Théorique, 13 rue Pierre et Marie Curie, F-75005, Paris, France 2) Institut de Biologie Physico-Chimique-Fondation Edmond de Rotschild, PSL Research University, Paris, France 3) Institut de Pharmacologie et de Biologie Structurale IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France Abstract Molecular visualisation is fundamental in the current scientific literature, textbooks and dissemination materials, forming an essential support for presenting results, reasoning on and formulating hypotheses related to molecular structure. Visual exploration has become easily accessible on a broad variety of platforms thanks to advanced software tools that render a great service to the scientific community. These tools are often developed across disciplines bridging computer science, biology and chemistry. Here we first describe a few Swiss Army knives geared towards protein visualisation for everyday use with an existing large user base, then focus on more specialised tools for peculiar needs that are not yet as broadly known. Our selection is by no means exhaustive, but reflects a diverse snapshot of scenarios that we consider informative for the reader. We end with an account of future trends and perspectives. Keywords Molecular Graphics, Protein visualization, Software tools, Virtual reality Introduction Many parts of science rely on a visualization-driven cycle of experimentation, reasoning, conjecture and validation, even more so in relation with structural biology and biophysics. Molecular visualization (1) in particular is now broadly used in many contexts, with the purpose of illustration in the scientific literature or the aim to gain insight about primary research data. -

Interdomain Zinc Site on Human Albumin SPECIAL FEATURE
Interdomain zinc site on human albumin SPECIAL FEATURE Alan J. Stewart*, Claudia A. Blindauer*, Stephen Berezenko†, Darrell Sleep†, and Peter J. Sadler*‡ *School of Chemistry, University of Edinburgh, West Mains Road, Edinburgh EH9 3JJ, United Kingdom; and †Delta Biotechnology Ltd., Castle Court, Castle Boulevard, Nottingham NG7 1FD, United Kingdom Edited by Jack Halpern, University of Chicago, Chicago, IL, and approved December 20, 2002 (received for review October 30, 2002) Albumin is the major transport protein in blood for Zn2؉, a metal binds to the thiolate sulfur at Cys-34 (23). CD studies suggest ion required for physiological processes and recruited by various that the major Zn2ϩ site is also a secondary (weaker) binding drugs and toxins. However, the Zn2؉-binding site(s) on albumin is site for Cu2ϩ and Ni2ϩ (17). Early 113Cd NMR experiments ϩ ill-defined. We have analyzed the 18 x-ray crystal structures of hu- on BSA demonstrated the existence of two Cd2 -binding sites man albumin in the PDB and identified a potential five-coordinate (18), A and B. Competition experiments on bovine and HSAs ϩ Zn site at the interface of domains I and II consisting of N ligands (18, 19, 24) have shown that site A binds Zn2 more strongly ϩ from His-67 and His-247 and O ligands from Asn-99, Asp-249, and than Cd2 . NMR peaks A (130 ppm in phosphate buffer) and 113 H2O, which are the same amino acid ligands as those in the zinc B (24 ppm) were also observed when Cd was added to in- enzymes calcineurin, endonucleotidase, and purple acid phospha- tact human blood serum, although peak A was shifted down- tase.