6339, AKR1B10, Human Recombinant
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Enzyme DHRS7
Toward the identification of a function of the “orphan” enzyme DHRS7 Inauguraldissertation zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Selene Araya, aus Lugano, Tessin Basel, 2018 Originaldokument gespeichert auf dem Dokumentenserver der Universität Basel edoc.unibas.ch Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät auf Antrag von Prof. Dr. Alex Odermatt (Fakultätsverantwortlicher) und Prof. Dr. Michael Arand (Korreferent) Basel, den 26.6.2018 ________________________ Dekan Prof. Dr. Martin Spiess I. List of Abbreviations 3α/βAdiol 3α/β-Androstanediol (5α-Androstane-3α/β,17β-diol) 3α/βHSD 3α/β-hydroxysteroid dehydrogenase 17β-HSD 17β-Hydroxysteroid Dehydrogenase 17αOHProg 17α-Hydroxyprogesterone 20α/βOHProg 20α/β-Hydroxyprogesterone 17α,20α/βdiOHProg 20α/βdihydroxyprogesterone ADT Androgen deprivation therapy ANOVA Analysis of variance AR Androgen Receptor AKR Aldo-Keto Reductase ATCC American Type Culture Collection CAM Cell Adhesion Molecule CYP Cytochrome P450 CBR1 Carbonyl reductase 1 CRPC Castration resistant prostate cancer Ct-value Cycle threshold-value DHRS7 (B/C) Dehydrogenase/Reductase Short Chain Dehydrogenase Family Member 7 (B/C) DHEA Dehydroepiandrosterone DHP Dehydroprogesterone DHT 5α-Dihydrotestosterone DMEM Dulbecco's Modified Eagle's Medium DMSO Dimethyl Sulfoxide DTT Dithiothreitol E1 Estrone E2 Estradiol ECM Extracellular Membrane EDTA Ethylenediaminetetraacetic acid EMT Epithelial-mesenchymal transition ER Endoplasmic Reticulum ERα/β Estrogen Receptor α/β FBS Fetal Bovine Serum 3 FDR False discovery rate FGF Fibroblast growth factor HEPES 4-(2-Hydroxyethyl)-1-Piperazineethanesulfonic Acid HMDB Human Metabolome Database HPLC High Performance Liquid Chromatography HSD Hydroxysteroid Dehydrogenase IC50 Half-Maximal Inhibitory Concentration LNCaP Lymph node carcinoma of the prostate mRNA Messenger Ribonucleic Acid n.d. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

The Identification of Human Aldo-Keto Reductase AKR7A2 As a Novel
Li et al. Cellular & Molecular Biology Letters (2016) 21:25 Cellular & Molecular DOI 10.1186/s11658-016-0026-9 Biology Letters SHORTCOMMUNICATION Open Access The identification of human aldo-keto reductase AKR7A2 as a novel cytoglobin- binding partner Xin Li, Shanshan Zou, Zhen Li, Gaotai Cai, Bohong Chen, Ping Wang and Wenqi Dong* * Correspondence: [email protected] Abstract Department of Biopharmaceutics, School of Laboratory Medicine and Cytoglobin (CYGB), a member of the globin family, is thought to protect cells from Biotechnology, Southern Medical reactive oxygen and nitrogen species and deal with hypoxic conditions and University, 1838 North Guangzhou oxidative stress. However, its molecular mechanisms of action are not clearly Avenue, Guangzhou 510515, China understood. Through immunoprecipitation combined with a two-dimensional electrophoresis–mass spectrometry assay, we identified a CYGB interactor: aldo-keto reductase family 7 member A2 (AKR7A2). The interaction was further confirmed using yeast two-hybrid and co-immunoprecipitation assays. Our results show that AKR7A2 physically interacts with CYGB. Keywords: CYGB, AKR7A2, Protein-protein interactions, Yeast two-hybrid assay, Co-immunoprecipitation, 2-DE, Oxidative stress Introduction Cytoglobin (CYGB), which is a member of the globin family, was discovered more than a decade ago in a proteomic screen of fibrotic liver [1]. It was originally named STAP (stellate activating protein). Human CYGB is a 190-amino acid, 21-kDa protein [2], encoded by a single copy gene mapped at the 17q25.3 chromosomal segment [3]. It has a compact helical conformation, giving it the ability to bind to heme, which allows reversible binding of gaseous, diatomic molecules, including oxygen (O2), nitric oxide (NO) and carbon monoxide (CO), just like hemoglobin (Hb), myoglobin (Mb) and neuroglobin (Ngb) [4]. -

AKR7A2 (NM 003689) Human Tagged ORF Clone Lentiviral Particle Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC214673L4V AKR7A2 (NM_003689) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: AKR7A2 (NM_003689) Human Tagged ORF Clone Lentiviral Particle Symbol: AKR7A2 Synonyms: AFAR; AFAR1; AFB1-AR1; AKR7 Vector: pLenti-C-mGFP-P2A-Puro (PS100093) ACCN: NM_003689 ORF Size: 1077 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC214673). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_003689.2 RefSeq Size: 1377 bp RefSeq ORF: 1080 bp Locus ID: 8574 UniProt ID: O43488, V9HWA2 Domains: aldo_ket_red Protein Families: Druggable Genome MW: 39.4 kDa This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 AKR7A2 (NM_003689) Human Tagged ORF Clone Lentiviral Particle – RC214673L4V Gene Summary: The protein encoded by this gene belongs to the aldo/keto reductase (AKR) superfamily and AKR7 family, which are involved in the detoxification of aldehydes and ketones. -

Reactive Carbonyls and Oxidative Stress: Potential for Therapeutic Intervention ⁎ Elizabeth M
Pharmacology & Therapeutics 115 (2007) 13–24 www.elsevier.com/locate/pharmthera Associate editor: R.M. Wadsworth Reactive carbonyls and oxidative stress: Potential for therapeutic intervention ⁎ Elizabeth M. Ellis Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, 204 George Street, Glasgow, G1 1XW, United Kingdom Abstract Reactive aldehydes and ketones are produced as a result of oxidative stress in several disease processes. Considerable evidence is now accumulating that these reactive carbonyl products are also involved in the progression of diseases, including neurodegenerative disorders, diabetes, atherosclerosis, diabetic complications, reperfusion after ischemic injury, hypertension, and inflammation. To counter carbonyl stress, cells possess enzymes that can decrease aldehyde load. These enzymes include aldehyde dehydrogenases (ALDH), aldo-keto reductases (AKR), carbonyl reductase (CBR), and glutathione S-transferases (GST). Some of these enzymes are inducible by chemoprotective compounds via Nrf2/ ARE- or AhR/XRE-dependent mechanisms. This review describes the metabolism of reactive carbonyls and discusses the potential for manipulating levels of carbonyl-metabolizing enzymes through chemical intervention. © 2007 Elsevier Inc. All rights reserved. Keywords: Aldehyde metabolism; Oxidative stress; Chemoprotection Contents 1. Introduction ............................................. 14 2. Production of reactive carbonyls in oxidant-exposed cells . .................... 14 2.1. Carbonyls produced -
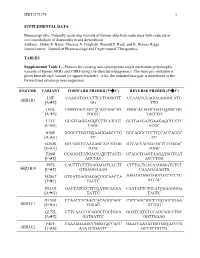
Data Supplement
JPET#173179 1 SUPPLEMENTAL DATA Manuscript title: Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin Authors: Onkar S. Bains, Thomas A. Grigliatti, Ronald E. Reid, and K. Wayne Riggs Journal name: Journal of Pharmacology and Experimental Therapeutics TABLES Supplemental Table 1—Primers for creating non-synonymous single nucleotide polymorphic variants of human AKRs and CBR4 using site-directed mutagenesis. The base pair mutation is given beneath each variant [in square brackets]. Also, the mutated base pair is underlined in the forward and reverse primer sequences. ENZYME VARIANT FORWARD PRIMER (5'3') REVERSE PRIMER (5'3') I15F CAAGATGCCCTTCCTGGGGTT CCAACCCCAGGAAGGGCATC AKR1B1 [AT] GG TTG H42L CGGGTACCGCCTCATCGACTG GGGCACAGTCGATGAGGCGG [AT] TGCCC TACCCG L73V GCGTGAGGAGGTCTTCATCGT GCTGACGATGAAGACCTCCTC [CG] CAGC ACGC K90E GGGCCTGGTGGAAGGAGCCTG GGCAGGCTCCTTCCACCAGGC [AG] CC CC G204S GCCAGTCCAAAAGCATCGTGG GTCACCACGATGCTTTTGGAC [GA] TGAC TGGC T288I CCAGGATATGACCATCTTACTC GTAGCTGAGTAAGATGGTCAT [CT] AGCTAC ATCCTGG P87S CACTTTCTTTGAGAGATCCCTT CTTTCCTCACAAGGGATCTCT AKR1B10 [CT] GTGAGGAAAG CAAAGAAAGTG M286T GTGATGAGGAGACGGCAACCA GAGTATGGTTGCCGTCTCCTC [TC] TACTC ATCAC N313D GACTATCCCTTCGATGCAGAA CAATATTCTGCATCGAAGGGA [AG] TATTG TAGTC R170H CCAACTTCAACCACAGGCAGC CTCCAGCTGCCTGTGGTTGAA AKR1C1 [GA] TGGAG GTTGG Q172L CTTCAACCGCAGGCTGCTGGA GGATCATCTCCAGCAGCCTGC [AT] GATGATCC GGTTGAAG F46Y CAATAGAAGCCGGGTACCACC GAATCAATATGGTGGTACCCG AKR1C2 [TA] ATATTTGATTC GCTTCTATTG JPET#173179 -

Computational Inferences of Mutations Driving Mesenchymal Differentiation in Glioblastoma
Computational Inferences of Mutations Driving Mesenchymal Differentiation in Glioblastoma James Chen Submitted in partial fulfillment of the requirements for the Doctor of Philosophy Degree in the Graduate School of Arts and Sciences Columbia University 2013 ! 2013 James Chen All rights reserved ABSTRACT Computational Inferences of Mutations Driving Mesenchymal Differentiation in Glioblastoma James Chen This dissertation reviews the development and implementation of integrative, systems biology methods designed to parse driver mutations from high- throughput array data derived from human patients. The analysis of vast amounts of genomic and genetic data in the context of complex human genetic diseases such as Glioblastoma is a daunting task. Mutations exist by the hundreds, if not thousands, and only an unknown handful will contribute to the disease in a significant way. The goal of this project was to develop novel computational methods to identify candidate mutations from these data that drive the molecular differentiation of glioblastoma into the mesenchymal subtype, the most aggressive, poorest-prognosis tumors associated with glioblastoma. TABLE OF CONTENTS CHAPTER 1… Introduction and Background 1 Glioblastoma and the Mesenchymal Subtype 3 Systems Biology and Master Regulators 9 Thesis Project: Genetics and Genomics 20 CHAPTER 2… TCGA Data Processing 23 CHAPTER 3… DIGGIn Part 1 – Selecting f-CNVs 33 Mutual Information 40 Application and Analysis 45 CHAPTER 4… DIGGIn Part 2 – Selecting drivers 52 CHAPTER 5… KLHL9 Manuscript 63 Methods 90 CHAPTER 5a… Revisions work-in-progress 105 CHAPTER 6… Discussion 109 APPENDICES… 132 APPEND01 – TCGA classifications 133 APPEND02 – GBM f-CNV list 136 APPEND03 – MES f-CNV candidate drivers 152 APPEND04 – Scripts 149 APPEND05 – Manuscript Figures and Legends 175 APPEND06 – Manuscript Supplemental Materials 185 i ACKNOWLEDGEMENTS I would like to thank the Califano Lab and my mentor, Andrea Califano, for their intellectual and motivational support during my stay in their lab. -

Table 3. PDB Representation of Gene Families A. H. Sapiens
Table 3. PDB representation of gene families A. H. -

Interindividual Variability in the Cardiac Expression of Anthracycline Reductases in Donors with and Without Down Syndrome
Pharm Res DOI 10.1007/s11095-013-1267-1 RESEARCH PAPER Interindividual Variability in the Cardiac Expression of Anthracycline Reductases in Donors With and Without Down Syndrome Adolfo Quiñones-Lombraña & Daniel Ferguson & Rachael Hageman Blair & James L. Kalabus & Almedina Redzematovic & Javier G. Blanco Received: 16 September 2013 /Accepted: 9 December 2013 # Springer Science+Business Media New York 2014 ABSTRACT CBR1 rs9024 genotype status impacts on cardiac CBR1 expres- Purpose The intracardiac synthesis of anthracycline alcohol sion in non-DS hearts. metabolites (e.g., daunorubicinol) contributes to the patho- Conclusions CBR1, AKR1A1, and AKR7A2 protein levels point genesis of anthracycline-related cardiotoxicity. Cancer pa- to be important determinants for predicting the synthesis of tients with Down syndrome (DS) are at increased risk for cardiotoxic daunorubicinol in heart. anthracycline-related cardiotoxicity. We profiled the expression of anthracycline metabolizing enzymes in hearts from donors with- KEY WORDS Aldo-keto reductases . Anthracycline-related and without- DS. cardiotoxicity . Anthracyclines . Carbonyl reductases . Down Methods Cardiac expression of CBR1, CBR3, AKR1A1, syndrome AKR1C3 and AKR7A2 was examined by quantitative real time PCR, quantitative immunoblotting, and enzyme activity assays ABBREVIATIONS using daunorubicin. The CBR1 polymorphism rs9024 was inves- aCGH Array comparative genomic hybridization tigated by allelic discrimination with fluorescent probes. The ACTB Actin B contribution of CBRs/AKRs proteins to daunorubicin reductase AKR1A1 Aldo-keto reductase family 1, member A1 activity was examined by multiple linear regression. AKR1C3 Aldo-keto reductase family 1, member C3 Results CBR1 was the most abundant transcript (average relative AKR7A2 Aldo-keto reductase family 7, member A2 expression; DS: 81%, non-DS: 58%), and AKR7A2 was the most AKRs Aldo-keto reductases abundant protein (average relative expression; DS: 38%, non- CBR1 Carbonyl reductase 1 DS: 35%). -

Molecular Mechanism and Metabolic Function of the S
MOLECULAR MECHANISM AND METABOLIC FUNCTION OF THE S- NITROSO-COENZYME A REDUCTASE AKR1A1 by COLIN T. STOMBERSKI Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation Advisor: Jonathan S. Stamler Department of Biochemistry CASE WESTERN RESERVE UNIVERSITY May, 2019 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the dissertation of COLIN T. STOMBERSKI candidate for the degree of Doctor of Philosophy*. Committee Chair Focco van den Akker Committee Members Jonathan Stamler George Dubyak Mukesh Jain Hung-Ying Kao 03-22-2019 *We also certify that written approval has been obtained for any proprietary material contained therein TABLE OF CONTENTS Table of Contents ………………………………………………………………………… i List of Tables ……………………………………………………………………………. v List of Figures ………………………………………………………………………….. vi List of Abbreviations …………………………………………………………………… ix Acknowledgements …………………………………………………………………….. xi Abstract ………………………………………………………………………………….. 1 Foundation and Experimental Framework ……………………………………………….. 3 Chapter 1: Protein S-nitrosylation: Determinants of specificity and enzymatic regulation of S-nitrosothiol-based signaling …………………………………………….. 5 1.1 Introduction …………………………………………………………………. 6 1.2 S-nitrosothiol specificity …………………………………………………….. 8 1.2.1 Acid-base and hydrophobic motifs ……………………………… 9 1.2.2 Interaction with nitric oxide synthases ………………………….. 13 1.3 S-nitrosothiol stability and reactivity ……………………………………….. 15 1.3.1 RSNO bond chemistry ………………………………………….. 16 1.3.2 Protein SNO—thiol reaction bias ………………………………. 18 1.3.3 SNO sites do not overlap S-oxidation sites …………………….. 19 1.4 Enzymatic denitrosylation ………………………………………………….. 20 1.4.1 The thioredoxin system ………………………………………… 21 1.4.2 LMW-SNO reductases …………………………………………. 23 1.4.3 The GSNO reductase system …………………………………… 24 1.4.4 GSNOR in physiology and pathophysiology …………………… 26 i 1.4.5 The SNO-CoA reductase system ……………………………….. 31 1.5 Specificity in denitrosylation ………………………………………………. -

Genetic Testing for Developmental Disabilities, Intellectual Disability, and Autism Spectrum Disorder Technical Brief Number 23
Technical Brief Number 23 Genetic Testing for Developmental Disabilities, Intellectual Disability, and Autism Spectrum Disorder Technical Brief Number 23 Genetic Testing for Developmental Disabilities, Intellectual Disability, and Autism Spectrum Disorder Prepared for: Agency for Healthcare Research and Quality U.S. Department of Health and Human Services 540 Gaither Road Rockville, MD 20850 www.ahrq.gov Contract No. 290-2012-00011-I Prepared by: ECRI Institute–Penn Medicine Evidence-based Practice Center Plymouth Meeting, PA Investigators: Fang Sun, M.D., Ph.D. Jeff Oristaglio, Ph.D. Susan E. Levy, M.D., M.P.H. Hakon Hakonarson, M.D., Ph.D. Nancy Sullivan, B.A. Joann Fontanarosa, Ph.D. Karen M. Schoelles, M.D., M.S., FACP AHRQ Publication No. 15-EHC024-EF June 2015 This report is based on research conducted by the ECRI–Penn Medicine AHRQ Evidence-based Practice Center (EPC) under contract to the Agency for Healthcare Research and Quality (AHRQ), Rockville, MD (290-2012-00011-I). The findings and conclusions in this document are those of the authors, who are responsible for its contents; the findings and conclusions do not necessarily represent the views of AHRQ. Therefore, no statement in this report should be construed as an official position of AHRQ or of the U.S. Department of Health and Human Services. The information in this report is intended to help health care decisionmakers—patients and clinicians, health system leaders, and policymakers, among others—make well-informed decisions and thereby improve the quality of health care services. This report is not intended to be a substitute for the application of clinical judgment. -

Review Multiplicity of Mammalian Reductases for Xenobiotic Carbonyl Compounds
Drug Metab. Pharmacokinet. 21 (1): 1–18 (2006). Review Multiplicity of Mammalian Reductases for Xenobiotic Carbonyl Compounds Toshiyuki MATSUNAGA, Shinichi SHINTANI and Akira HARA* Laboratory of Biochemistry, Gifu Pharmaceutical University, Gifu, Japan Full text of this paper is available at http://www.jstage.jst.go.jp/browse/dmpk Summary: A variety of carbonyl compounds are present in foods, environmental pollutants, and drugs. These xenobiotic carbonyl compounds are metabolized into the corresponding alcohols by many mammalian NAD(P)H-dependent reductases, which belong to the short-chain dehydrogenaseWreductase (SDR) and aldo-keto reductase superfamilies. Recent genomic analysis, cDNA isolation and characteriza- tion of the recombinant enzymes suggested that, in humans, the six members of each of the two superfamilies, i.e., total of 12 enzymes, are involved in the reductive metabolism of xenobiotic carbonyl compounds. They comprise three types of carbonyl reductase, dehydrogenaseWreductase (SDR family) member 4, 11b-hydroxysteroid dehydrogenase type 1, L-xylulose reductase, two types of a‰atoxin B1 aldehyde reductase, 20a-hydroxysteroid dehydrogenase, and three types of 3a-hydroxysteroid dehydrogenase. Accumulating data on the human enzymes provide new insights into their roles in cellular and molecular reactions including xenobiotic metabolism. On the other hand, mice and rats lack the gene for a protein corresponding to human 3a-hydroxysteroid dehydrogenase type 3, but instead possess additional ˆve or six genes encoding proteins that are structurally related to human hydroxy- steroid dehydrogenases. Characterization of the additional enzymes suggested their involvement in species-speciˆc biological events and species diŠerences in the metabolism of xenobiotic carbonyl compounds. Key words: Carbonyl reduction; short-chain dehydrogenaseWreductase superfamily; aldo-keto reductase superfamily; carbonyl reductase; hydroxysteroid dehydrogenase; species diŠerence alcohols by NADPH-dependent reductases with broad Introduction substrate speciˆcity.