Reactive Carbonyls and Oxidative Stress: Potential for Therapeutic Intervention ⁎ Elizabeth M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Enzyme DHRS7
Toward the identification of a function of the “orphan” enzyme DHRS7 Inauguraldissertation zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Selene Araya, aus Lugano, Tessin Basel, 2018 Originaldokument gespeichert auf dem Dokumentenserver der Universität Basel edoc.unibas.ch Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät auf Antrag von Prof. Dr. Alex Odermatt (Fakultätsverantwortlicher) und Prof. Dr. Michael Arand (Korreferent) Basel, den 26.6.2018 ________________________ Dekan Prof. Dr. Martin Spiess I. List of Abbreviations 3α/βAdiol 3α/β-Androstanediol (5α-Androstane-3α/β,17β-diol) 3α/βHSD 3α/β-hydroxysteroid dehydrogenase 17β-HSD 17β-Hydroxysteroid Dehydrogenase 17αOHProg 17α-Hydroxyprogesterone 20α/βOHProg 20α/β-Hydroxyprogesterone 17α,20α/βdiOHProg 20α/βdihydroxyprogesterone ADT Androgen deprivation therapy ANOVA Analysis of variance AR Androgen Receptor AKR Aldo-Keto Reductase ATCC American Type Culture Collection CAM Cell Adhesion Molecule CYP Cytochrome P450 CBR1 Carbonyl reductase 1 CRPC Castration resistant prostate cancer Ct-value Cycle threshold-value DHRS7 (B/C) Dehydrogenase/Reductase Short Chain Dehydrogenase Family Member 7 (B/C) DHEA Dehydroepiandrosterone DHP Dehydroprogesterone DHT 5α-Dihydrotestosterone DMEM Dulbecco's Modified Eagle's Medium DMSO Dimethyl Sulfoxide DTT Dithiothreitol E1 Estrone E2 Estradiol ECM Extracellular Membrane EDTA Ethylenediaminetetraacetic acid EMT Epithelial-mesenchymal transition ER Endoplasmic Reticulum ERα/β Estrogen Receptor α/β FBS Fetal Bovine Serum 3 FDR False discovery rate FGF Fibroblast growth factor HEPES 4-(2-Hydroxyethyl)-1-Piperazineethanesulfonic Acid HMDB Human Metabolome Database HPLC High Performance Liquid Chromatography HSD Hydroxysteroid Dehydrogenase IC50 Half-Maximal Inhibitory Concentration LNCaP Lymph node carcinoma of the prostate mRNA Messenger Ribonucleic Acid n.d. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

The Identification of Human Aldo-Keto Reductase AKR7A2 As a Novel
Li et al. Cellular & Molecular Biology Letters (2016) 21:25 Cellular & Molecular DOI 10.1186/s11658-016-0026-9 Biology Letters SHORTCOMMUNICATION Open Access The identification of human aldo-keto reductase AKR7A2 as a novel cytoglobin- binding partner Xin Li, Shanshan Zou, Zhen Li, Gaotai Cai, Bohong Chen, Ping Wang and Wenqi Dong* * Correspondence: [email protected] Abstract Department of Biopharmaceutics, School of Laboratory Medicine and Cytoglobin (CYGB), a member of the globin family, is thought to protect cells from Biotechnology, Southern Medical reactive oxygen and nitrogen species and deal with hypoxic conditions and University, 1838 North Guangzhou oxidative stress. However, its molecular mechanisms of action are not clearly Avenue, Guangzhou 510515, China understood. Through immunoprecipitation combined with a two-dimensional electrophoresis–mass spectrometry assay, we identified a CYGB interactor: aldo-keto reductase family 7 member A2 (AKR7A2). The interaction was further confirmed using yeast two-hybrid and co-immunoprecipitation assays. Our results show that AKR7A2 physically interacts with CYGB. Keywords: CYGB, AKR7A2, Protein-protein interactions, Yeast two-hybrid assay, Co-immunoprecipitation, 2-DE, Oxidative stress Introduction Cytoglobin (CYGB), which is a member of the globin family, was discovered more than a decade ago in a proteomic screen of fibrotic liver [1]. It was originally named STAP (stellate activating protein). Human CYGB is a 190-amino acid, 21-kDa protein [2], encoded by a single copy gene mapped at the 17q25.3 chromosomal segment [3]. It has a compact helical conformation, giving it the ability to bind to heme, which allows reversible binding of gaseous, diatomic molecules, including oxygen (O2), nitric oxide (NO) and carbon monoxide (CO), just like hemoglobin (Hb), myoglobin (Mb) and neuroglobin (Ngb) [4]. -

AKR7A2 (NM 003689) Human Tagged ORF Clone Lentiviral Particle Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC214673L4V AKR7A2 (NM_003689) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: AKR7A2 (NM_003689) Human Tagged ORF Clone Lentiviral Particle Symbol: AKR7A2 Synonyms: AFAR; AFAR1; AFB1-AR1; AKR7 Vector: pLenti-C-mGFP-P2A-Puro (PS100093) ACCN: NM_003689 ORF Size: 1077 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC214673). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_003689.2 RefSeq Size: 1377 bp RefSeq ORF: 1080 bp Locus ID: 8574 UniProt ID: O43488, V9HWA2 Domains: aldo_ket_red Protein Families: Druggable Genome MW: 39.4 kDa This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 AKR7A2 (NM_003689) Human Tagged ORF Clone Lentiviral Particle – RC214673L4V Gene Summary: The protein encoded by this gene belongs to the aldo/keto reductase (AKR) superfamily and AKR7 family, which are involved in the detoxification of aldehydes and ketones. -

In Vivo Effect of Oracin on Doxorubicin Reduction, Biodistribution and Efficacy in Ehrlich Tumor Bearing Mice
Pharmacological Reports Copyright © 2013 2013, 65, 445452 by Institute of Pharmacology ISSN 1734-1140 Polish Academy of Sciences Invivo effectoforacinondoxorubicinreduction, biodistributionandefficacyinEhrlichtumor bearingmice VeronikaHanušová1,PavelTomšík2,LenkaKriesfalusyová3, AlenaPakostová1,IvaBoušová1,LenkaSkálová1 1 DepartmentofBiochemicalSciences,CharlesUniversityinPrague,FacultyofPharmacy,Heyrovského1203, HradecKrálové,CZ-50005,CzechRepublic 2 DepartmentofMedicalBiochemistry,CharlesUniversityinPrague,FacultyofMedicine, Šimkova570, HradecKrálové,CZ-50038,CzechRepublic 3 Radio-IsotopeLaboratory,CharlesUniversityinPrague,FacultyofMedicine, Šimkova570,HradecKrálové, CZ-50038,CzechRepublic Correspondence: LenkaSkálová,e-mail:[email protected] Abstract: Background: The limitation of carbonyl reduction represents one possible way to increase the effectiveness of anthracycline doxo- rubicin (DOX) in cancer cells and decrease its toxicity in normal cells. In vitro, isoquinoline derivative oracin (ORC) inhibited DOX reduction and increased the antiproliferative effect of DOX in MCF-7 breast cancer cells. Moreover, ORC significantly decreases DOX toxicity in non-cancerous MCF-10A breast cells and in hepatocytes. The present study was designed to test in mice the in vivo effect of ORC on plasma and tissue concentrations of DOX and its main metabolite DOXOL. The effect of ORC on DOX efficacy in micebearingsolidEhrlichtumors(EST)wasalsostudied. Methods: DOX and DOX + ORC combinations were iv administered to healthy mice. Blood samples, livers -

Biochemical and Pharmacological Characterization of Cytochrome B5 Reductase As a Potential Novel Therapeutic Target in Candida A
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2011 Biochemical and Pharmacological Characterization of Cytochrome b5 Reductase as a Potential Novel Therapeutic Target in Candida albicans Mary Jolene Patricia Holloway University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Biochemistry Commons, Microbiology Commons, and the Molecular Biology Commons Scholar Commons Citation Holloway, Mary Jolene Patricia, "Biochemical and Pharmacological Characterization of Cytochrome b5 Reductase as a Potential Novel Therapeutic Target in Candida albicans" (2011). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/3730 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Biochemical and Pharmacological Characterization of Cytochrome b5 Reductase as a Potential Novel Therapeutic Target in Candida albicans by Mary Jolene Holloway A dissertation written in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Molecular Medicine College of Medicine University of South Florida Major Professor: Andreas Seyfang, Ph.D. Co-Major Professor: Robert Deschenes, Ph.D. Michael J. Barber, D.Phil Gloria Ferreira, Ph.D. Alberto van Olphen, Ph.D. Date of Approval: November 14, 2011 Keywords: Fungal drug resistance, protein biochemistry and structure, ergosterol biosynthesis, yeast knockouts, oxidoreductases Copyright © 2011, Mary Jolene Holloway i DEDICATION This work is dedicated to my mother, Theresa Holloway, for being my coach and friend. -

Anti-Oxidant Pathogenesis of High-Grade Glioma DISSERTATION
Anti-Oxidant Pathogenesis of High-Grade Glioma DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Ji Eun Song, M.S. Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2015 Dissertation Committee: Dr. Chang-Hyuk Kwon, Advisor Dr. Balveen Kaur, Co-advisor Dr. Vincenzo Coppola Dr. Thomas Ludwig Copyright by Ji Eun Song 2015 Abstract High-grade glioma (HGG) is the most aggressive primary brain malignancies, and is incurable despite the best combination of current cancer therapies. A median patient survival of glioblastoma (GBM, the most aggressive grade 4 glioma) is only 14.6 months (Stupp et al., 2005). Therefore, innovative and more effective therapy for HGG is urgently needed. It has been known that dysregulated reactive oxygen species (ROS) signaling is associated with many human diseases, including cancers. Oxidative stress by excessive accumulation of ROS has been known to promote carcinogenesis through both genetic and epigenetic modifications (Ziech, Franco, Pappa, & Panayiotidis, 2011). Expressions of anti-oxidant proteins are reportedly increased by ROS- induced oxidative stress (Polytarchou, Pfau, Hatziapostolou, & Tsichlis, 2008). Because excessive oxidative stress can cause cellular senescence and apoptosis, it appears that tumor cells overexpress anti-oxidant proteins as a defense mechanism against elevated ROS. Therefore, targeting a predominant anti-oxidant protein could be an effective strategy for treating tumors. Peroxiredoxin 4 (PRDX4) is an ROS-scavenging enzyme and facilitates proper protein folding in the endoplasmic reticulum (ER). We reported that PRDX4 levels ii were highly increased in a majority of human HGGs as well as in a mouse model of HGG. -
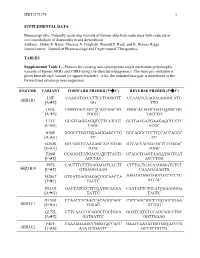
Data Supplement
JPET#173179 1 SUPPLEMENTAL DATA Manuscript title: Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin Authors: Onkar S. Bains, Thomas A. Grigliatti, Ronald E. Reid, and K. Wayne Riggs Journal name: Journal of Pharmacology and Experimental Therapeutics TABLES Supplemental Table 1—Primers for creating non-synonymous single nucleotide polymorphic variants of human AKRs and CBR4 using site-directed mutagenesis. The base pair mutation is given beneath each variant [in square brackets]. Also, the mutated base pair is underlined in the forward and reverse primer sequences. ENZYME VARIANT FORWARD PRIMER (5'3') REVERSE PRIMER (5'3') I15F CAAGATGCCCTTCCTGGGGTT CCAACCCCAGGAAGGGCATC AKR1B1 [AT] GG TTG H42L CGGGTACCGCCTCATCGACTG GGGCACAGTCGATGAGGCGG [AT] TGCCC TACCCG L73V GCGTGAGGAGGTCTTCATCGT GCTGACGATGAAGACCTCCTC [CG] CAGC ACGC K90E GGGCCTGGTGGAAGGAGCCTG GGCAGGCTCCTTCCACCAGGC [AG] CC CC G204S GCCAGTCCAAAAGCATCGTGG GTCACCACGATGCTTTTGGAC [GA] TGAC TGGC T288I CCAGGATATGACCATCTTACTC GTAGCTGAGTAAGATGGTCAT [CT] AGCTAC ATCCTGG P87S CACTTTCTTTGAGAGATCCCTT CTTTCCTCACAAGGGATCTCT AKR1B10 [CT] GTGAGGAAAG CAAAGAAAGTG M286T GTGATGAGGAGACGGCAACCA GAGTATGGTTGCCGTCTCCTC [TC] TACTC ATCAC N313D GACTATCCCTTCGATGCAGAA CAATATTCTGCATCGAAGGGA [AG] TATTG TAGTC R170H CCAACTTCAACCACAGGCAGC CTCCAGCTGCCTGTGGTTGAA AKR1C1 [GA] TGGAG GTTGG Q172L CTTCAACCGCAGGCTGCTGGA GGATCATCTCCAGCAGCCTGC [AT] GATGATCC GGTTGAAG F46Y CAATAGAAGCCGGGTACCACC GAATCAATATGGTGGTACCCG AKR1C2 [TA] ATATTTGATTC GCTTCTATTG JPET#173179 -

Chromosome 21 Leading Edge Gene Set
Chromosome 21 Leading Edge Gene Set Genes from chr21q22 that are part of the GSEA leading edge set identifying differences between trisomic and euploid samples. Multiple probe set IDs corresponding to a single gene symbol are combined as part of the GSEA analysis. Gene Symbol Probe Set IDs Gene Title 203865_s_at, 207999_s_at, 209979_at, adenosine deaminase, RNA-specific, B1 ADARB1 234539_at, 234799_at (RED1 homolog rat) UDP-Gal:betaGlcNAc beta 1,3- B3GALT5 206947_at galactosyltransferase, polypeptide 5 BACE2 217867_x_at, 222446_s_at beta-site APP-cleaving enzyme 2 1553227_s_at, 214820_at, 219280_at, 225446_at, 231860_at, 231960_at, bromodomain and WD repeat domain BRWD1 244622_at containing 1 C21orf121 240809_at chromosome 21 open reading frame 121 C21orf130 240068_at chromosome 21 open reading frame 130 C21orf22 1560881_a_at chromosome 21 open reading frame 22 C21orf29 1552570_at, 1555048_at_at, 1555049_at chromosome 21 open reading frame 29 C21orf33 202217_at, 210667_s_at chromosome 21 open reading frame 33 C21orf45 219004_s_at, 228597_at, 229671_s_at chromosome 21 open reading frame 45 C21orf51 1554430_at, 1554432_x_at, 228239_at chromosome 21 open reading frame 51 C21orf56 223360_at chromosome 21 open reading frame 56 C21orf59 218123_at, 244369_at chromosome 21 open reading frame 59 C21orf66 1555125_at, 218515_at, 221158_at chromosome 21 open reading frame 66 C21orf7 221211_s_at chromosome 21 open reading frame 7 C21orf77 220826_at chromosome 21 open reading frame 77 C21orf84 239968_at, 240589_at chromosome 21 open reading frame 84 -

Computational Inferences of Mutations Driving Mesenchymal Differentiation in Glioblastoma
Computational Inferences of Mutations Driving Mesenchymal Differentiation in Glioblastoma James Chen Submitted in partial fulfillment of the requirements for the Doctor of Philosophy Degree in the Graduate School of Arts and Sciences Columbia University 2013 ! 2013 James Chen All rights reserved ABSTRACT Computational Inferences of Mutations Driving Mesenchymal Differentiation in Glioblastoma James Chen This dissertation reviews the development and implementation of integrative, systems biology methods designed to parse driver mutations from high- throughput array data derived from human patients. The analysis of vast amounts of genomic and genetic data in the context of complex human genetic diseases such as Glioblastoma is a daunting task. Mutations exist by the hundreds, if not thousands, and only an unknown handful will contribute to the disease in a significant way. The goal of this project was to develop novel computational methods to identify candidate mutations from these data that drive the molecular differentiation of glioblastoma into the mesenchymal subtype, the most aggressive, poorest-prognosis tumors associated with glioblastoma. TABLE OF CONTENTS CHAPTER 1… Introduction and Background 1 Glioblastoma and the Mesenchymal Subtype 3 Systems Biology and Master Regulators 9 Thesis Project: Genetics and Genomics 20 CHAPTER 2… TCGA Data Processing 23 CHAPTER 3… DIGGIn Part 1 – Selecting f-CNVs 33 Mutual Information 40 Application and Analysis 45 CHAPTER 4… DIGGIn Part 2 – Selecting drivers 52 CHAPTER 5… KLHL9 Manuscript 63 Methods 90 CHAPTER 5a… Revisions work-in-progress 105 CHAPTER 6… Discussion 109 APPENDICES… 132 APPEND01 – TCGA classifications 133 APPEND02 – GBM f-CNV list 136 APPEND03 – MES f-CNV candidate drivers 152 APPEND04 – Scripts 149 APPEND05 – Manuscript Figures and Legends 175 APPEND06 – Manuscript Supplemental Materials 185 i ACKNOWLEDGEMENTS I would like to thank the Califano Lab and my mentor, Andrea Califano, for their intellectual and motivational support during my stay in their lab. -

RACK1 Promotes Hepatocellular Carcinoma Cell Survival Via CBR1 by Suppressing TNF-Α-Induced ROS Generation
ONCOLOGY LETTERS 12: 5303-5308, 2016 RACK1 promotes hepatocellular carcinoma cell survival via CBR1 by suppressing TNF-α-induced ROS generation SILEI ZHOU1,2*, HUANLING CAO1,2*, YAWEI ZHAO1,2, XINYING LI1, JIYAN ZHANG1, CHUNMEI HOU1, YUANFANG MA2 and QINGYANG WANG1 1Department of Molecular Immunology, Institute of Basic Medical Sciences, Beijing 100850; 2Laboratory of Cellular and Molecular Immunology, Henan University, Kaifeng, Henan 475004, P.R. China Received April 24, 2015; Accepted September 9, 2016 DOI: 10.3892/ol.2016.5339 Abstract. It has been reported that intracellular accumulation This process is regulated by a number of intracellular signaling of reactive oxygen species (ROS) has a significant role in tumor pathways, including c-jun N-terminal kinase (JNK) and IκB necrosis factor (TNF)-α-induced cell apoptosis and necrosis; kinase (IKK), as well as reactive oxygen species (ROS) (4,5). however, the key molecules regulating ROS generation remain Extensive studies have indicated that reduced levels of oxidant to be elucidated. The present study reports that knockdown stress and ROS promote malignant transformation and onco- of endogenous receptor for activated C kinase 1 (RACK1) genic growth in hepatocellular carcinoma (HCC) cells (6-9). increases the intracellular ROS level following TNF-α or However, the key molecules regulating ROS in HCC remain H2O2 stimulation in human hepatocellular carcinoma (HCC) to be elucidated. It has been reported that scaffolding protein cells, leading to promotion of cell death. Carbonyl reductase 1 receptor for activated C kinase 1 (RACK1) enhances JNK (CBR1), a ubiquitous nicotinamide adenine dinucleotide activation in HCC, leading to promotion of the malignant phosphate-dependent enzyme, is reported to protect cells from growth of HCC (10).